scATAC-seq analysis
Here we will use scATAC-seq dataset “buenrostro2018” as an example to illustrate how SIMBA performs scATAC-seq analysis
[1]:
import os
import simba as si
si.__version__
[1]:
'1.0'
[2]:
workdir = 'result_simba_atacseq'
si.settings.set_workdir(workdir)
Saving results in: result_simba_atacseq
[3]:
si.settings.set_figure_params(dpi=80,
style='white',
fig_size=[5,5],
rc={'image.cmap': 'viridis'})
[4]:
# make plots prettier
from IPython.display import set_matplotlib_formats
set_matplotlib_formats('retina')
[ ]:
load example data
[6]:
adata_CP = si.datasets.atac_buenrostro2018()
atac_buenrostro2018.h5ad: 0.00B [00:00, ?B/s]
Downloading data ...
atac_buenrostro2018.h5ad: 145MB [01:45, 1.38MB/s]
Downloaded to result_simba_atacseq/data.
[7]:
adata_CP
[7]:
AnnData object with n_obs × n_vars = 2034 × 237450
obs: 'celltype'
var: 'chr', 'start', 'end'
[ ]:
preprocessing
[8]:
si.pp.filter_peaks(adata_CP,min_n_cells=3)
Before filtering:
2034 cells, 237450 peaks
Filter peaks based on min_n_cells
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
After filtering out low-expressed peaks:
2034 cells, 200167 peaks
[9]:
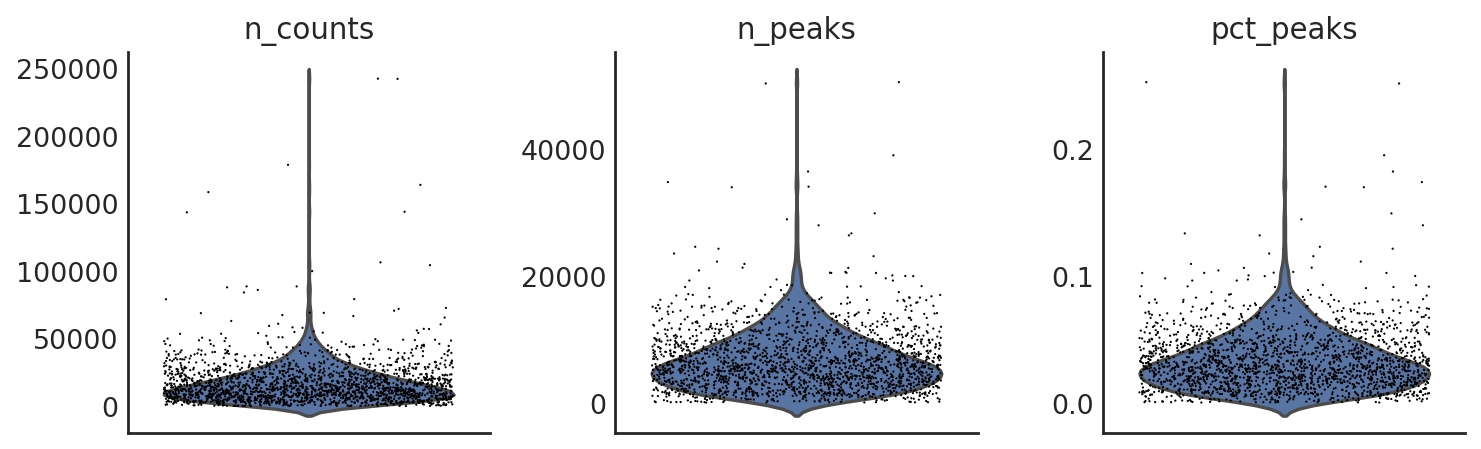
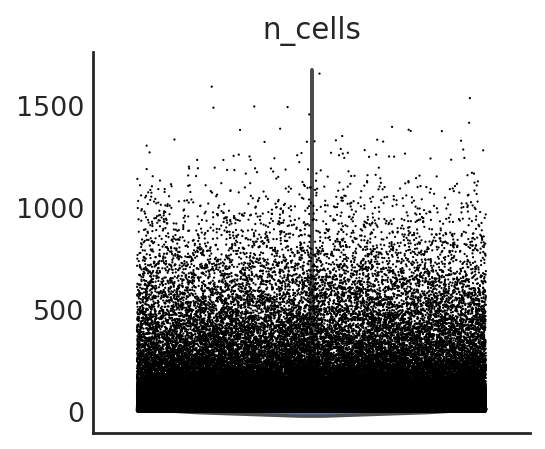
si.pp.cal_qc_atac(adata_CP)
different ways to visualize the QC metrics
[10]:
si.pl.violin(adata_CP,list_obs=['n_counts','n_peaks','pct_peaks'], list_var=['n_cells'],fig_size=(3,3))
[11]:
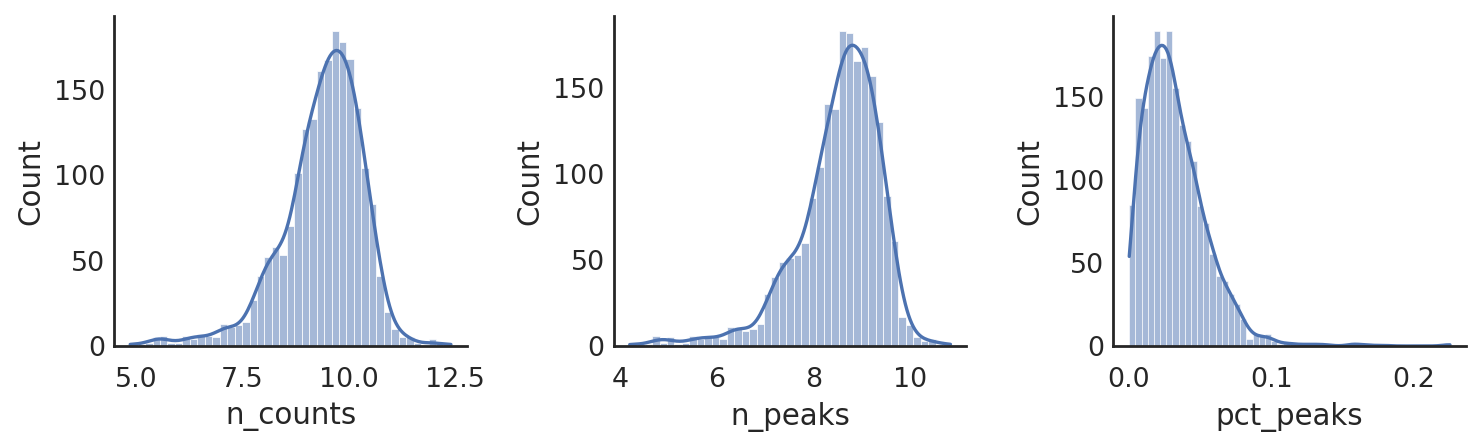
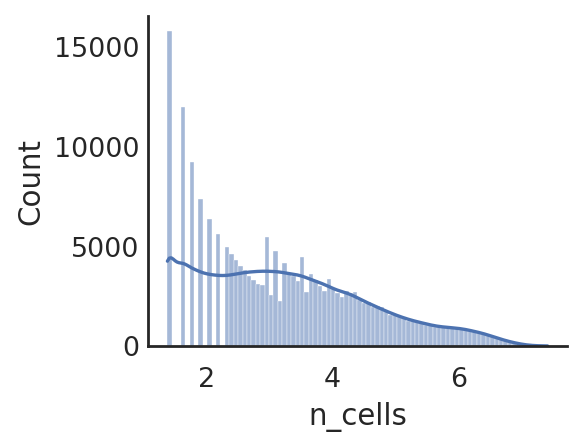
si.pl.hist(adata_CP,list_obs=['n_counts','n_peaks','pct_peaks'], log=True, list_var=['n_cells'],fig_size=(3,3))
[ ]:
Filter out cells if needed:
si.pp.filter_cells_atac(adata_CP,min_n_peaks=100)
[ ]:
select peaks (optional)
It will speed up the trainning process by only keeping peaks associated with top PCs
[12]:
si.pp.pca(adata_CP, n_components=50)
[13]:
si.pl.pca_variance_ratio(adata_CP, show_cutoff=True)
the number of selected PC is: 50
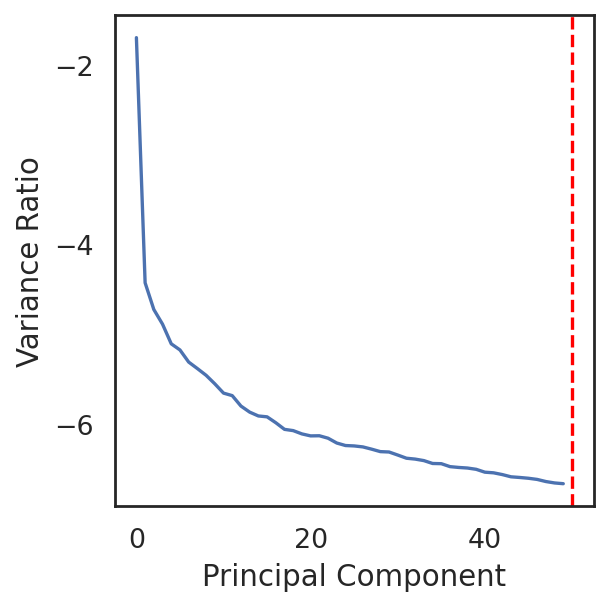
The number of selected PCs can be further reduced:
si.pp.select_pcs(adata_CP,n_pcs=30)
Select features associated with each of these PCs. The number of features for each PC will be automatically decided
[14]:
si.pp.select_pcs_features(adata_CP)
#features selected from PC 0: 33368
#features selected from PC 1: 33415
#features selected from PC 2: 33361
#features selected from PC 3: 33397
#features selected from PC 4: 33375
#features selected from PC 5: 33431
#features selected from PC 6: 33395
#features selected from PC 7: 33442
#features selected from PC 8: 33372
#features selected from PC 9: 33376
#features selected from PC 10: 33364
#features selected from PC 11: 33393
#features selected from PC 12: 33372
#features selected from PC 13: 33434
#features selected from PC 14: 33369
#features selected from PC 15: 33395
#features selected from PC 16: 33422
#features selected from PC 17: 33379
#features selected from PC 18: 33366
#features selected from PC 19: 33498
#features selected from PC 20: 33365
#features selected from PC 21: 33569
#features selected from PC 22: 33364
#features selected from PC 23: 33376
#features selected from PC 24: 33427
#features selected from PC 25: 33367
#features selected from PC 26: 33381
#features selected from PC 27: 33387
#features selected from PC 28: 33466
#features selected from PC 29: 33508
#features selected from PC 30: 33385
#features selected from PC 31: 33391
#features selected from PC 32: 33409
#features selected from PC 33: 33363
#features selected from PC 34: 33387
#features selected from PC 35: 33374
#features selected from PC 36: 33380
#features selected from PC 37: 33399
#features selected from PC 38: 33418
#features selected from PC 39: 33382
#features selected from PC 40: 33465
#features selected from PC 41: 33388
#features selected from PC 42: 33370
#features selected from PC 43: 33364
#features selected from PC 44: 33433
#features selected from PC 45: 33375
#features selected from PC 46: 33367
#features selected from PC 47: 33423
#features selected from PC 48: 33380
#features selected from PC 49: 33392
#features in total: 134535
[15]:
si.pl.pcs_features(adata_CP, fig_ncol=10)
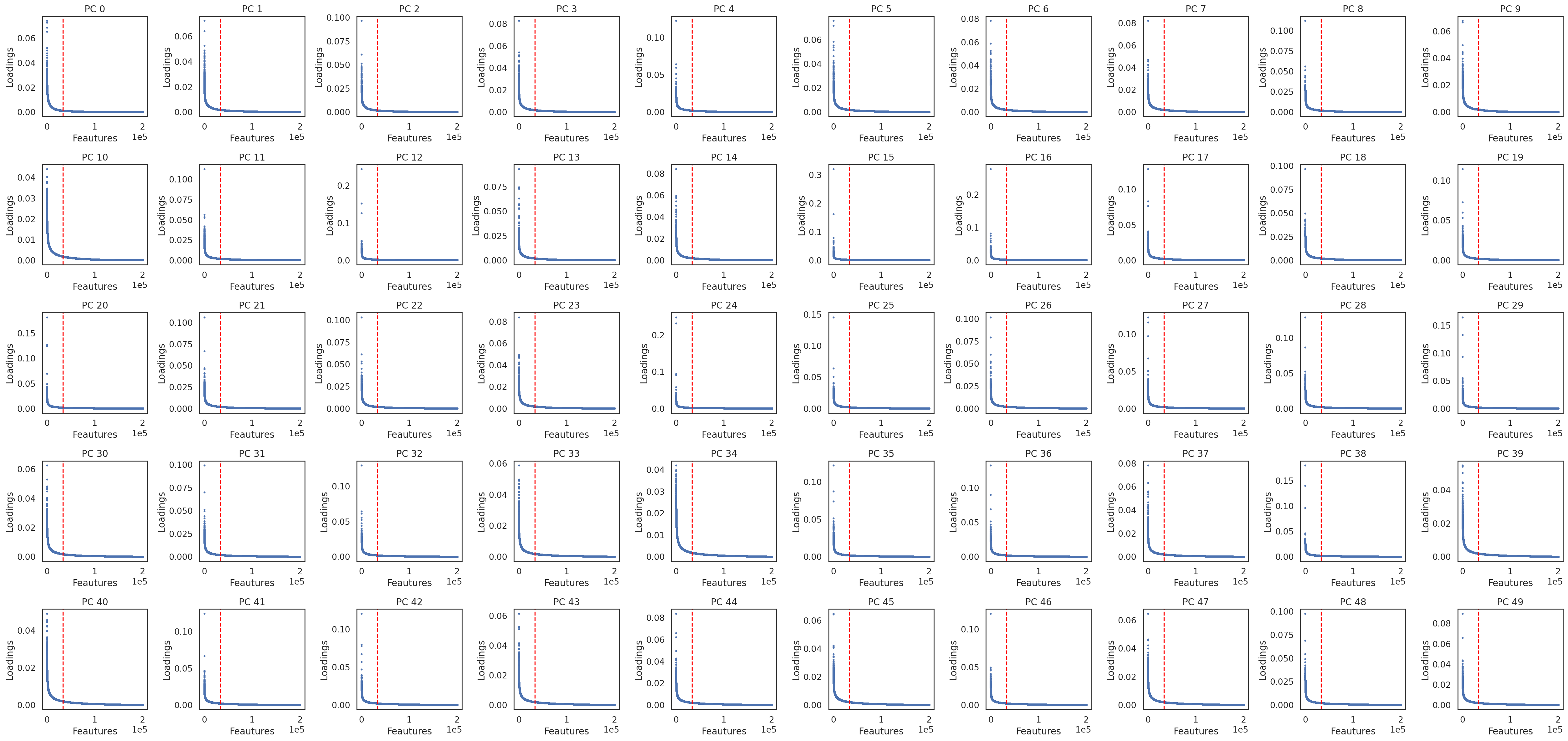
[ ]:
select DNA sequences (Optional)
DNA sequences such as TF motifs or k-mers can be also encoded into the graph if needed.
To scan peaks for kmers and motifs, first write the peak bed files.
[16]:
si.write_bed(adata_CP, use_top_pcs=True)
"peaks.bed" was written to "result_multiome_shareseq".
Then run the provided R script “scan_for_kmers_motifs.R” under the result directory (‘hg19.fa’ can be downloaded here)
[17]:
os.chdir(workdir)
[18]:
#! wget https://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/hg19.fa.gz
#! gunzip hg19.fa.gz
! wget https://raw.githubusercontent.com/pinellolab/simba/master/R_scripts/scan_for_kmers_motifs.R
--2021-11-09 17:40:40-- https://raw.githubusercontent.com/pinellolab/simba/master/R_scripts/scan_for_kmers_motifs.R
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.108.133, 185.199.109.133, 185.199.111.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.108.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 5767 (5.6K) [text/plain]
Saving to: ‘scan_for_kmers_motifs.R’
scan_for_kmers_moti 100%[===================>] 5.63K --.-KB/s in 0s
2021-11-09 17:40:40 (11.3 MB/s) - ‘scan_for_kmers_motifs.R’ saved [5767/5767]
Install the required packages in your enviroment with the following command:
$ conda install r-essentials r-optparse bioconductor-jaspar2020 bioconductor-biostrings bioconductor-tfbstools bioconductor-motifmatchr bioconductor-summarizedexperiment r-doparallel bioconductor-rhdf5 bioconductor-hdf5array
[19]:
%time
! Rscript scan_for_kmers_motifs.R -i peaks.bed -g hg19.fa -s 'Homo sapiens'
CPU times: user 7 µs, sys: 1 µs, total: 8 µs
Wall time: 14.8 µs
[1] "Converting .bed to .fasta ..."
[1] "Scanning for kmers ..."
[1] "Scanning for TF motifs ..."
[1] "Saving kmer matrix ..."
[1] "Saving motif matrix ..."
[1] "Finished."
[20]:
os.chdir("../")
[ ]:
[21]:
adata_PK = si.read_hdf(os.path.join(workdir,'output_kmers_motifs/freq_kmer.h5'),'mat')
adata_PM = si.read_hdf(os.path.join(workdir,'output_kmers_motifs/freq_motif.h5'),'mat')
# convert byte string to string
adata_PK.obs.index = [x.decode('utf-8') for x in adata_PK.obs.index]
adata_PK.var.index = [x.decode('utf-8') for x in adata_PK.var.index]
adata_PM.obs.index = [x.decode('utf-8') for x in adata_PM.obs.index]
adata_PM.var.index = [x.decode('utf-8') for x in adata_PM.var.index]
[22]:
adata_PK
[22]:
AnnData object with n_obs × n_vars = 134535 × 4096
[23]:
adata_PM
[23]:
AnnData object with n_obs × n_vars = 134535 × 633
[24]:
si.pp.binarize(adata_PK)
si.pp.binarize(adata_PM)
select kmers and motifs (optional)
[25]:
si.pp.pca(adata_PK, n_components=50)
si.pp.pca(adata_PM, n_components=50)
[26]:
si.pp.select_pcs_features(adata_PK, min_elbow=adata_PK.shape[1]/5, S=5)
si.pp.select_pcs_features(adata_PM, min_elbow=adata_PM.shape[1]/5, S=5)
#features selected from PC 0: 819
#features selected from PC 1: 828
#features selected from PC 2: 1491
#features selected from PC 3: 1495
#features selected from PC 4: 1981
#features selected from PC 5: 1710
#features selected from PC 6: 1277
#features selected from PC 7: 1877
#features selected from PC 8: 819
#features selected from PC 9: 1622
#features selected from PC 10: 1464
#features selected from PC 11: 1523
#features selected from PC 12: 1557
#features selected from PC 13: 1671
#features selected from PC 14: 1767
#features selected from PC 15: 1755
#features selected from PC 16: 2032
#features selected from PC 17: 1975
#features selected from PC 18: 819
#features selected from PC 19: 1843
#features selected from PC 20: 2072
#features selected from PC 21: 1766
#features selected from PC 22: 1711
#features selected from PC 23: 1861
#features selected from PC 24: 2002
#features selected from PC 25: 1891
#features selected from PC 26: 1820
#features selected from PC 27: 1886
#features selected from PC 28: 2082
#features selected from PC 29: 1446
#features selected from PC 30: 1557
#features selected from PC 31: 819
#features selected from PC 32: 2105
#features selected from PC 33: 1782
#features selected from PC 34: 1486
#features selected from PC 35: 1576
#features selected from PC 36: 1628
#features selected from PC 37: 1768
#features selected from PC 38: 1839
#features selected from PC 39: 819
#features selected from PC 40: 1833
#features selected from PC 41: 1496
#features selected from PC 42: 1329
#features selected from PC 43: 987
#features selected from PC 44: 1708
#features selected from PC 45: 1510
#features selected from PC 46: 1640
#features selected from PC 47: 1438
#features selected from PC 48: 2001
#features selected from PC 49: 1758
#features in total: 3601
#features selected from PC 0: 126
#features selected from PC 1: 189
#features selected from PC 2: 260
#features selected from PC 3: 270
#features selected from PC 4: 411
#features selected from PC 5: 339
#features selected from PC 6: 326
#features selected from PC 7: 312
#features selected from PC 8: 301
#features selected from PC 9: 274
#features selected from PC 10: 284
#features selected from PC 11: 185
#features selected from PC 12: 231
#features selected from PC 13: 270
#features selected from PC 14: 127
#features selected from PC 15: 296
#features selected from PC 16: 314
#features selected from PC 17: 270
#features selected from PC 18: 228
#features selected from PC 19: 340
#features selected from PC 20: 358
#features selected from PC 21: 262
#features selected from PC 22: 338
#features selected from PC 23: 265
#features selected from PC 24: 326
#features selected from PC 25: 308
#features selected from PC 26: 266
#features selected from PC 27: 286
#features selected from PC 28: 401
#features selected from PC 29: 343
#features selected from PC 30: 291
#features selected from PC 31: 228
#features selected from PC 32: 315
#features selected from PC 33: 324
#features selected from PC 34: 308
#features selected from PC 35: 313
#features selected from PC 36: 279
#features selected from PC 37: 288
#features selected from PC 38: 302
#features selected from PC 39: 310
#features selected from PC 40: 126
#features selected from PC 41: 375
#features selected from PC 42: 260
#features selected from PC 43: 362
#features selected from PC 44: 126
#features selected from PC 45: 265
#features selected from PC 46: 282
#features selected from PC 47: 320
#features selected from PC 48: 356
#features selected from PC 49: 238
#features in total: 601
[27]:
si.pl.pcs_features(adata_PK, fig_ncol=10)
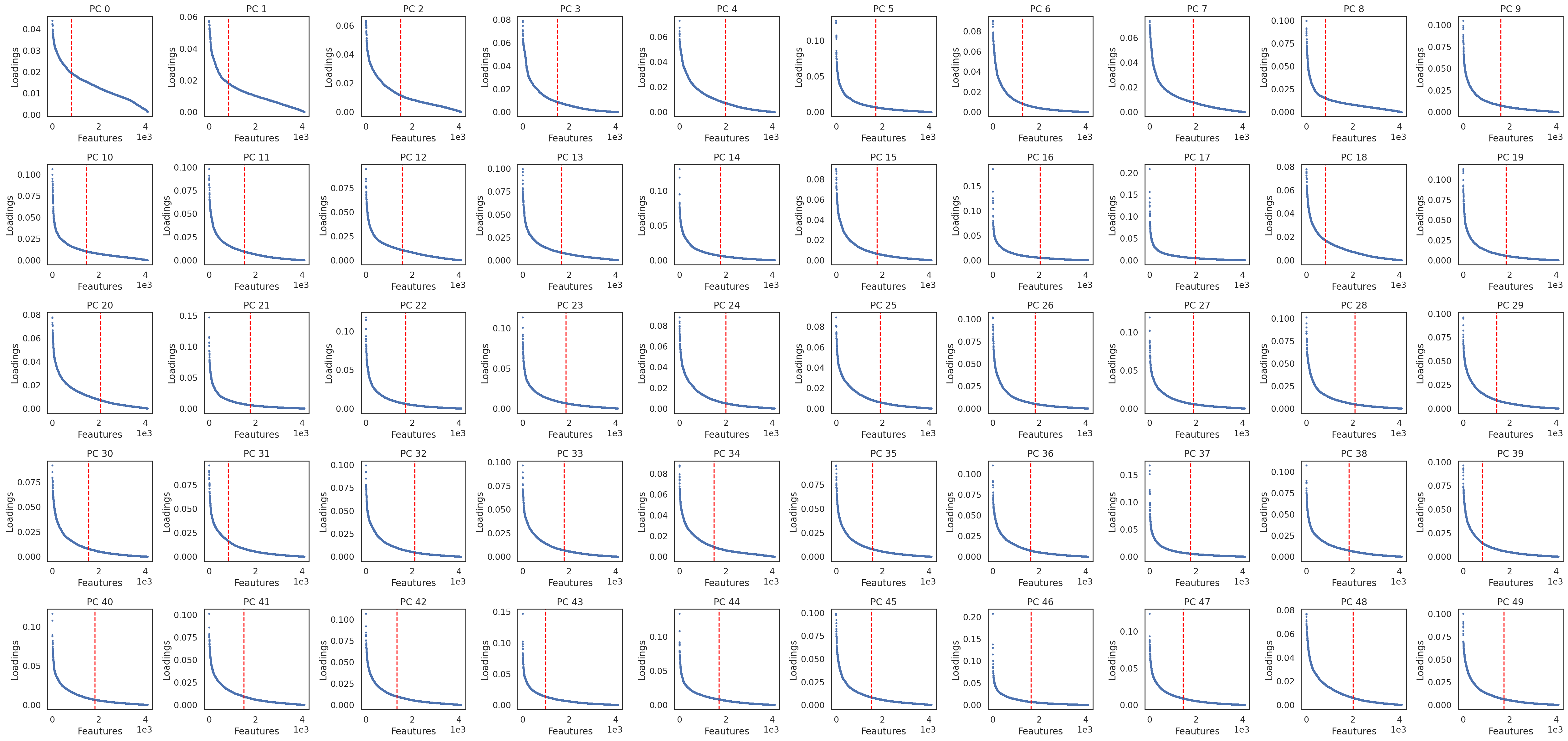
[28]:
si.pl.pcs_features(adata_PM, fig_ncol=10)
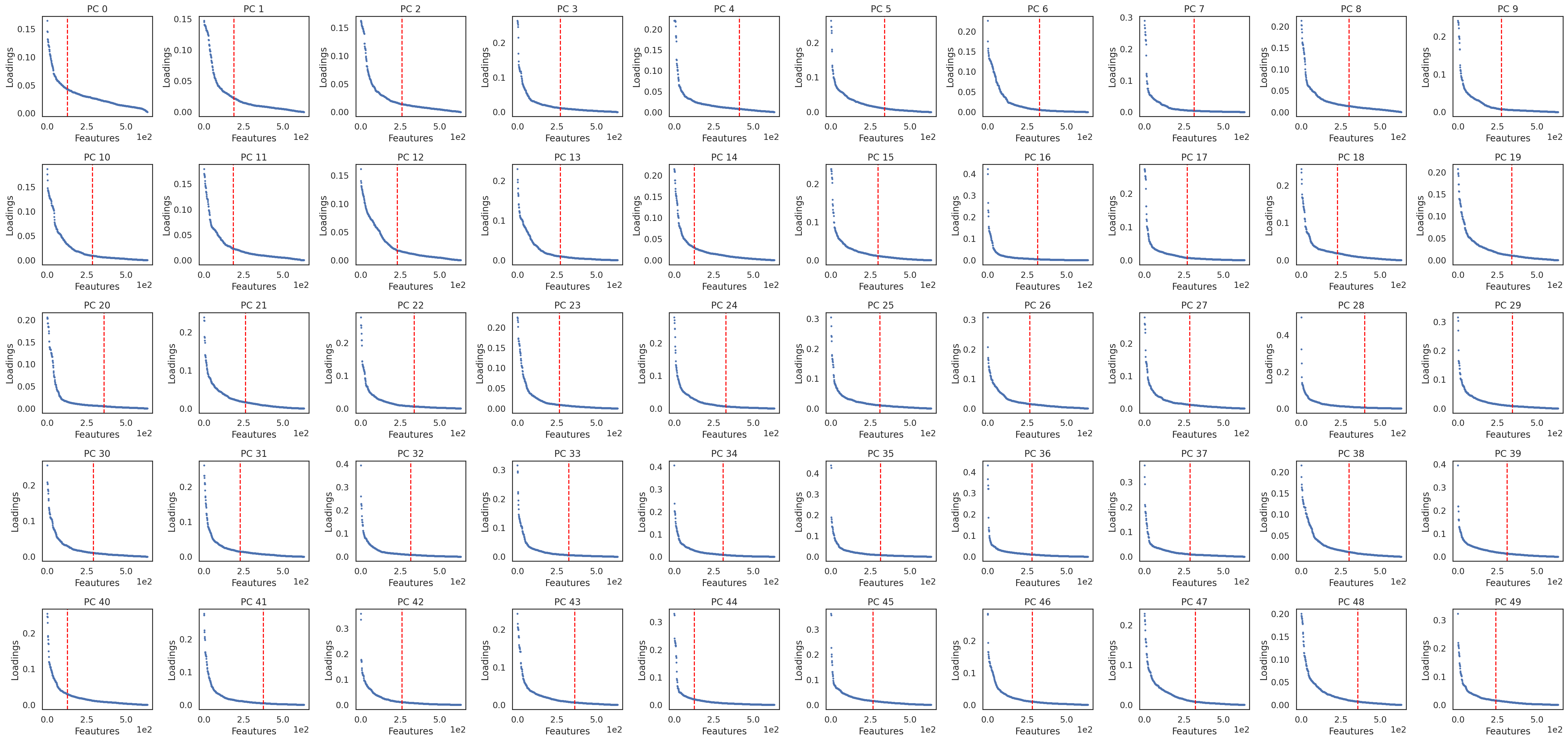
[ ]:
generate graph
[29]:
si.tl.gen_graph(list_CP=[adata_CP],
list_PK=[adata_PK],
list_PM=[adata_PM],
copy=False,
use_top_pcs=True,
dirname='graph0')
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
relation0: source: C, destination: P
#edges: 12977763
relation1: source: P, destination: M
#edges: 5749259
relation2: source: P, destination: K
#edges: 57893992
Total number of edges: 76621014
Writing graph file "pbg_graph.txt" to "result_simba_atacseq/pbg/graph0" ...
Finished.
[ ]:
PBG training
Before PBG training, let’s take a look at the parameters:
[30]:
si.settings.pbg_params
[30]:
{'entity_path': 'result_simba_atacseq/pbg/graph0/input/entity',
'edge_paths': ['result_simba_atacseq/pbg/graph0/input/edge'],
'checkpoint_path': '',
'entities': {'C': {'num_partitions': 1},
'P': {'num_partitions': 1},
'K': {'num_partitions': 1},
'M': {'num_partitions': 1}},
'relations': [{'name': 'r0',
'lhs': 'C',
'rhs': 'P',
'operator': 'none',
'weight': 1.0},
{'name': 'r1', 'lhs': 'P', 'rhs': 'M', 'operator': 'none', 'weight': 0.2},
{'name': 'r2', 'lhs': 'P', 'rhs': 'K', 'operator': 'none', 'weight': 0.02}],
'dynamic_relations': False,
'dimension': 50,
'global_emb': False,
'comparator': 'dot',
'num_epochs': 10,
'workers': 4,
'num_batch_negs': 50,
'num_uniform_negs': 50,
'loss_fn': 'softmax',
'lr': 0.1,
'early_stopping': False,
'regularization_coef': 0.0,
'wd': 0.0,
'wd_interval': 50,
'eval_fraction': 0.05,
'eval_num_batch_negs': 50,
'eval_num_uniform_negs': 50,
'checkpoint_preservation_interval': None}
[ ]:
If no parameters need to be adjusted, the training can be simply done with:
si.tl.pbg_train(auto_wd=True, save_wd=True, output='model')
[ ]:
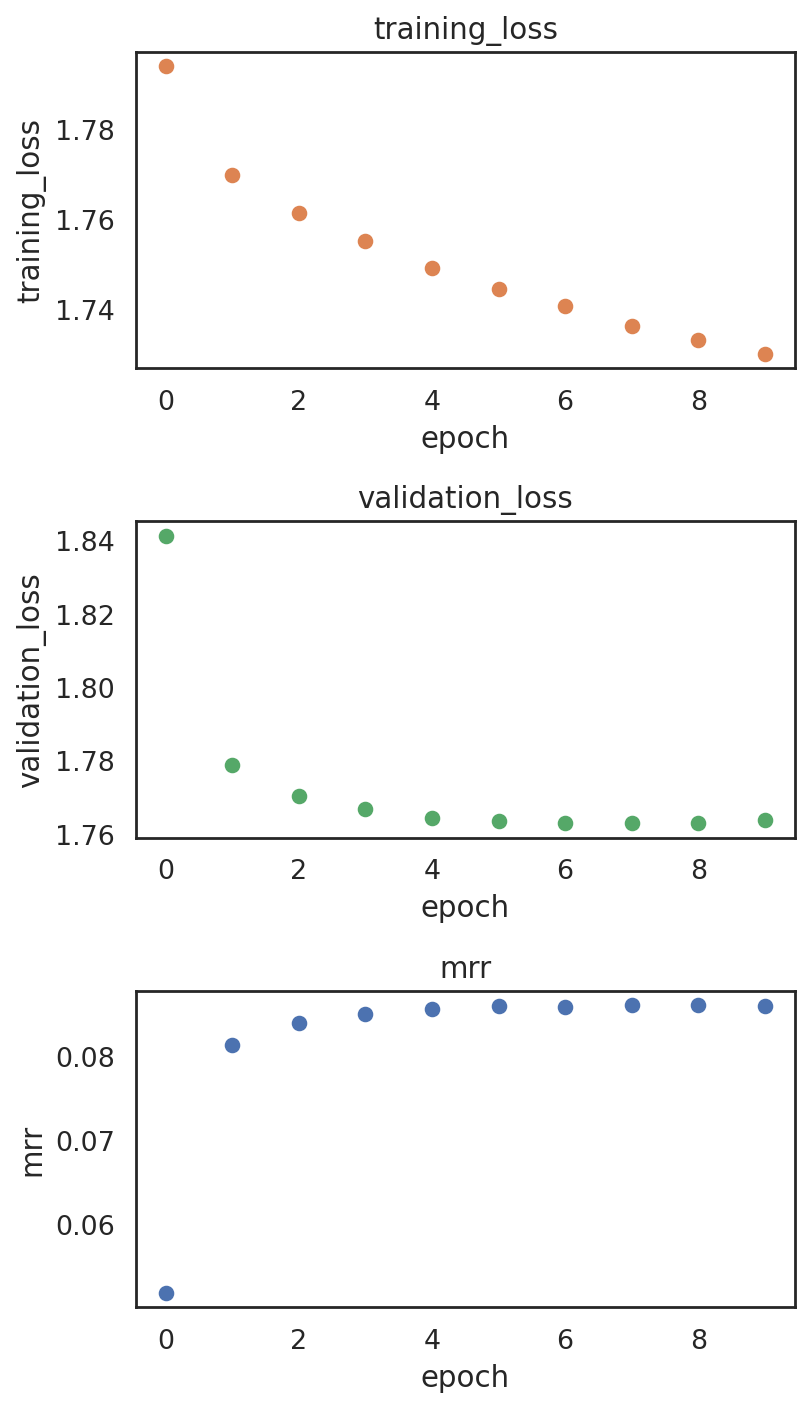
Here we show how to adjust training-related parameters if needed. In general, weight decay wd is the only parameter that might need to be adjusted based on the following pbg metric plots. However, in almost all the cases, the automatically decided wd (enabling it by setting auto_wd=True) works well.
E.g. we want to change the number of cpus workers:
[ ]:
[31]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
# dict_config['wd'] = 0.000309
dict_config['workers'] = 12
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model')
Auto-estimated weight decay is 0.000309
`.settings.pbg_params['wd']` has been updated to 0.000309
Converting input data ...
[2021-06-24 14:59:10.962921] Found some files that indicate that the input data has already been preprocessed, not doing it again.
[2021-06-24 14:59:10.963252] These files are in: result_simba_atacseq/pbg/graph0/input/entity, result_simba_atacseq/pbg/graph0/input/edge
Starting training ...
Finished
[ ]:
If
wdis specified by users instead of being automatically decided, then make sure to update it in simba setting:
si.settings.pbg_params = dict_config.copy()
[ ]:
The trained result can be loaded in with the following steps:
By default, it’s using the current training result stored in .setting.pbg_params
# load in graph ('graph0') info
si.load_graph_stats()
# load in model info for ('graph0')
si.load_pbg_config()
Users can also specify different pathss
# load in graph ('graph0') info
si.load_graph_stats(path='./result_simba_atacseq/pbg/graph0/')
# load in model info for ('graph0')
si.load_pbg_config(path='./result_simba_atacseq/pbg/graph0/model/')
[ ]:
plotting training metrics to make sure the model is not overfitting
[32]:
si.pl.pbg_metrics(fig_ncol=1)
[ ]:
post-training analysis
[33]:
palette_celltype={'HSC':"#00441B", 'MPP':"#46A040", 'LMPP':"#00AF99", 'CMP':"#FFC179",'CLP':"#98D9E9",
'MEP':"#F6313E", 'GMP':"#FFA300", 'pDC':"#C390D4", 'mono':"#FF5A00",'UNK':"#333333"}
[34]:
dict_adata = si.read_embedding()
[35]:
dict_adata
[35]:
{'C': AnnData object with n_obs × n_vars = 2034 × 50,
'K': AnnData object with n_obs × n_vars = 3601 × 50,
'P': AnnData object with n_obs × n_vars = 134535 × 50,
'M': AnnData object with n_obs × n_vars = 601 × 50}
[37]:
adata_C = dict_adata['C'] # embeddings for cells
adata_P = dict_adata['P'] # embeddings for peaks
adata_M = dict_adata['M'] # embeddings for motifs
adata_K = dict_adata['K'] # embeddings for kmers
[38]:
adata_C
[38]:
AnnData object with n_obs × n_vars = 2034 × 50
[39]:
adata_P
[39]:
AnnData object with n_obs × n_vars = 134535 × 50
[40]:
adata_M
[40]:
AnnData object with n_obs × n_vars = 601 × 50
[41]:
adata_K
[41]:
AnnData object with n_obs × n_vars = 3601 × 50
[ ]:
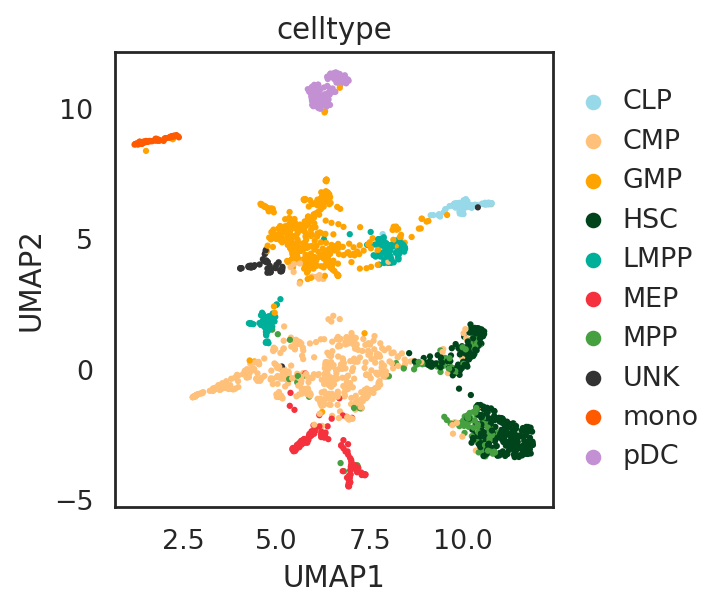
visualize embeddings of cells
[42]:
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CP[adata_C.obs_names,:].obs['celltype'].copy()
adata_C
[42]:
AnnData object with n_obs × n_vars = 2034 × 50
obs: 'celltype'
[43]:
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
[44]:
si.pl.umap(adata_C,color=['celltype'],
dict_palette={'celltype': palette_celltype},
fig_size=(4.5,4),
drawing_order='random')
[ ]:
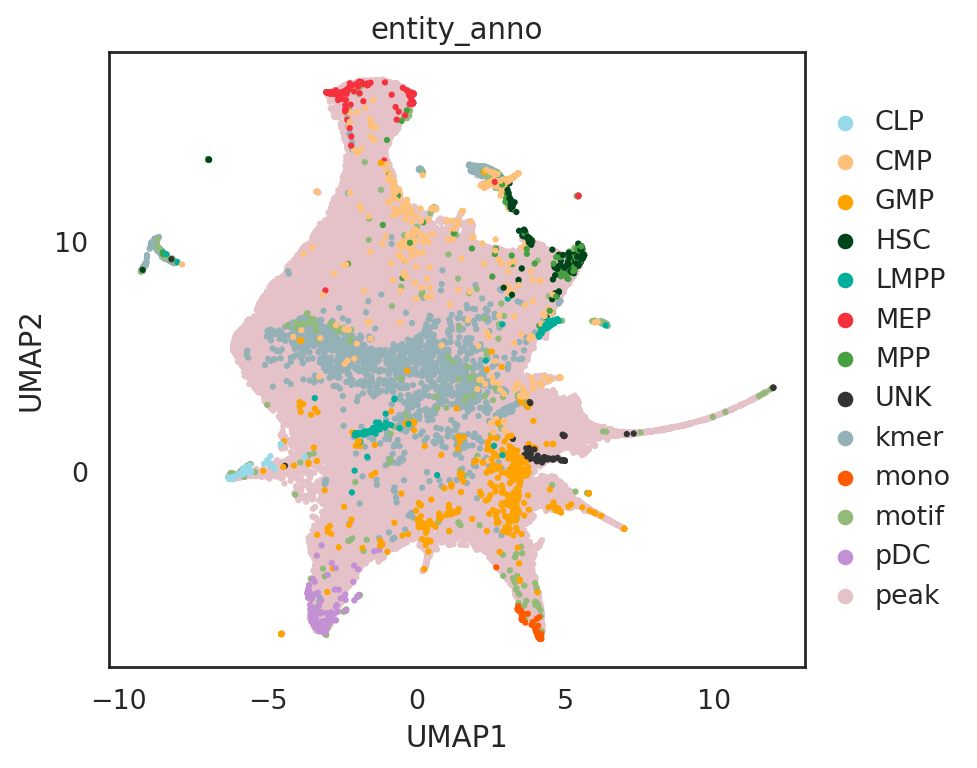
visualize embeddings of cells, motifs, kmers, and peaks
[45]:
adata_all = si.tl.embed(adata_ref=adata_C,
list_adata_query=[adata_M, adata_K, adata_P])
Performing softmax transformation for query data 0;
Performing softmax transformation for query data 1;
Performing softmax transformation for query data 2;
[46]:
## add annotations of all entities
adata_all.obs['entity_anno'] = ""
adata_all.obs.loc[adata_C.obs_names, 'entity_anno'] = adata_all.obs.loc[adata_C.obs_names, 'celltype'].tolist()
adata_all.obs.loc[adata_P.obs_names, 'entity_anno'] = 'peak'
adata_all.obs.loc[adata_K.obs_names, 'entity_anno'] = 'kmer'
adata_all.obs.loc[adata_M.obs_names, 'entity_anno'] = 'motif'
adata_all.obs.head()
[46]:
| celltype | id_dataset | entity_anno | |
|---|---|---|---|
| singles-BM0828-GMP-151027-21 | GMP | ref | GMP |
| singles-BM0828-MPP-frozen-151103-68 | MPP | ref | MPP |
| singles-160822-BM1137-CMP-LS-92 | CMP | ref | CMP |
| singles-PB1022-mono-160128-29 | mono | ref | mono |
| singles-160819-BM1137-CMP-LS-8 | CMP | ref | CMP |
[47]:
si.tl.umap(adata_all,n_neighbors=15,n_components=2)
[48]:
palette_entity_anno = palette_celltype.copy()
palette_entity_anno['peak'] = "#e5c2c8"
palette_entity_anno['kmer'] = "#94b1b7"
palette_entity_anno['motif'] = "#92ba79"
[49]:
si.pl.umap(adata_all[::-1,], #reverse the order of entities
color=['entity_anno'],
dict_palette={'entity_anno': palette_entity_anno},
fig_size=(6,5),
drawing_order='original',
show_texts=False)
Trying to set attribute `.uns` of view, copying.
[ ]:
[50]:
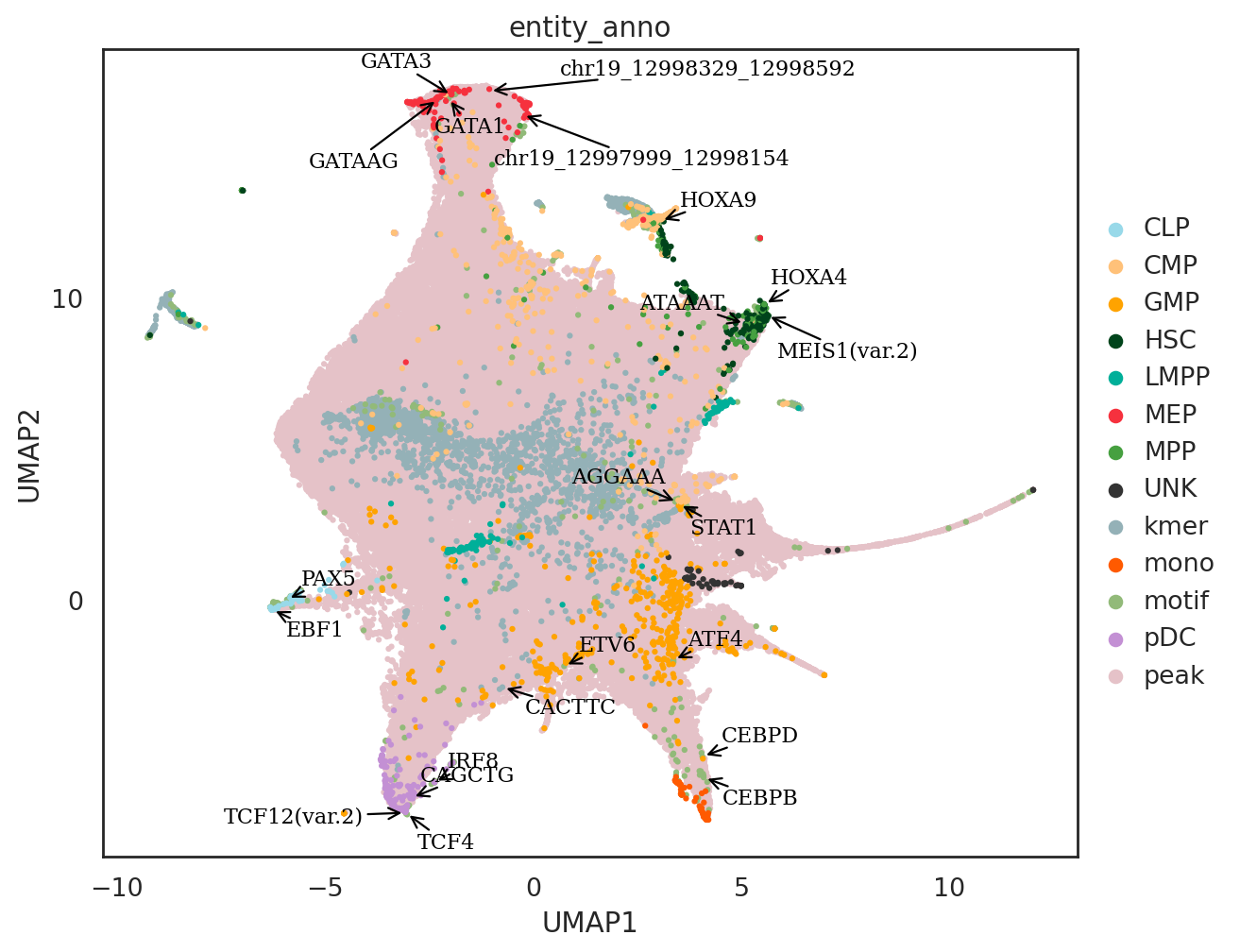
marker_tfs = ['PAX5', 'EBF1', # CLP
'TCF12(var.2)', 'IRF8', 'TCF4',#pDC
'CEBPB','CEBPD', #mono
'MEIS1(var.2)', 'HOXA9', 'HOXA4',#HSC
'GATA1', 'GATA3', #MEP
'ATF4','STAT1',#CMP
'ETV6', #GMP
]
marker_kmers = ['GATAAG', # GATA1
'CAGCTG', # TCF4/TCF12
'ATAAAT', # HOXA9
'AGGAAA', # STAT1
'CACTTC', #ETV6
]
marker_peaks = ['chr19_12997999_12998154', #KLF1
'chr19_12998329_12998592', #KLF1
]
[51]:
si.pl.umap(adata_all[::-1,],
color=['entity_anno'],
dict_palette={'entity_anno': palette_entity_anno},
drawing_order='original',
texts=marker_tfs + marker_kmers + marker_peaks,
text_expand=(1.5,2),
show_texts=True,
fig_size=(8,6.5))
Trying to set attribute `.uns` of view, copying.
[ ]:
SIMBA metrics
[52]:
# motifs
adata_cmp_CM = si.tl.compare_entities(adata_ref=adata_C,
adata_query=adata_M)
[53]:
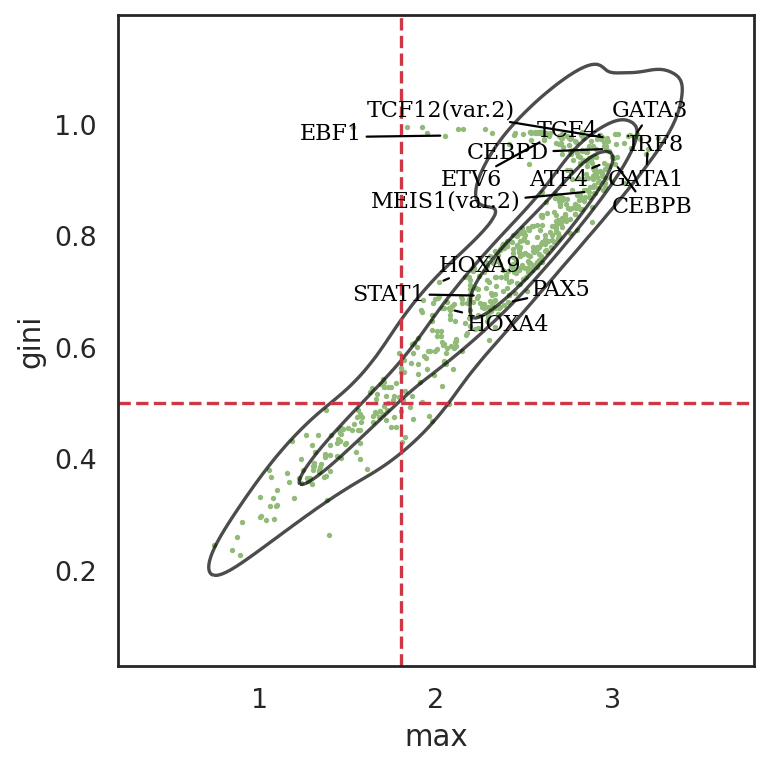
si.pl.entity_metrics(adata_cmp_CM,
x='max',
y='gini',
show_texts=True,
show_cutoff=True,
show_contour=True,
texts=marker_tfs,
text_expand=(1.5,1.5),
size=2,
cutoff_x=1.8,
cutoff_y=0.5,
c='#92ba79',
save_fig=False)
[ ]:
[54]:
# kmers
adata_cmp_CK = si.tl.compare_entities(adata_ref=adata_C,
adata_query=adata_K)
[55]:
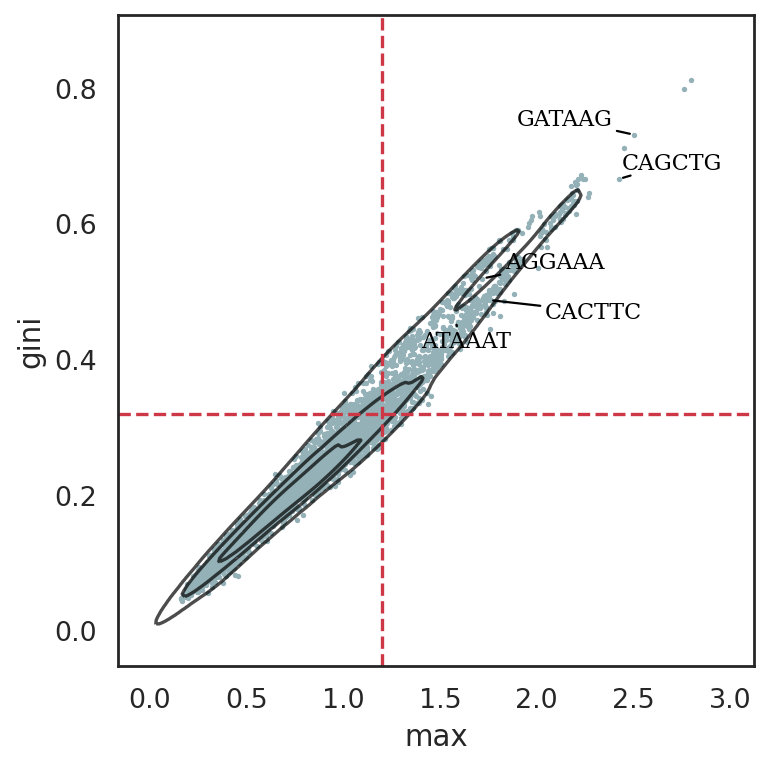
si.pl.entity_metrics(adata_cmp_CK,
x='max',
y='gini',
show_texts=True,
show_cutoff=True,
show_contour=True,
texts=marker_kmers,
text_expand=(1.5,1.5),
size=2,
cutoff_x=1.2,
cutoff_y=0.32,
c='#94b1b7',
save_fig=False)
[ ]:
SIMBA barcode plots
[56]:
adata_cmp_CM.obs['celltype'] = adata_CP.obs.loc[adata_cmp_CM.obs_names,'celltype']
adata_cmp_CK.obs['celltype'] = adata_CP.obs.loc[adata_cmp_CK.obs_names,'celltype']
[59]:
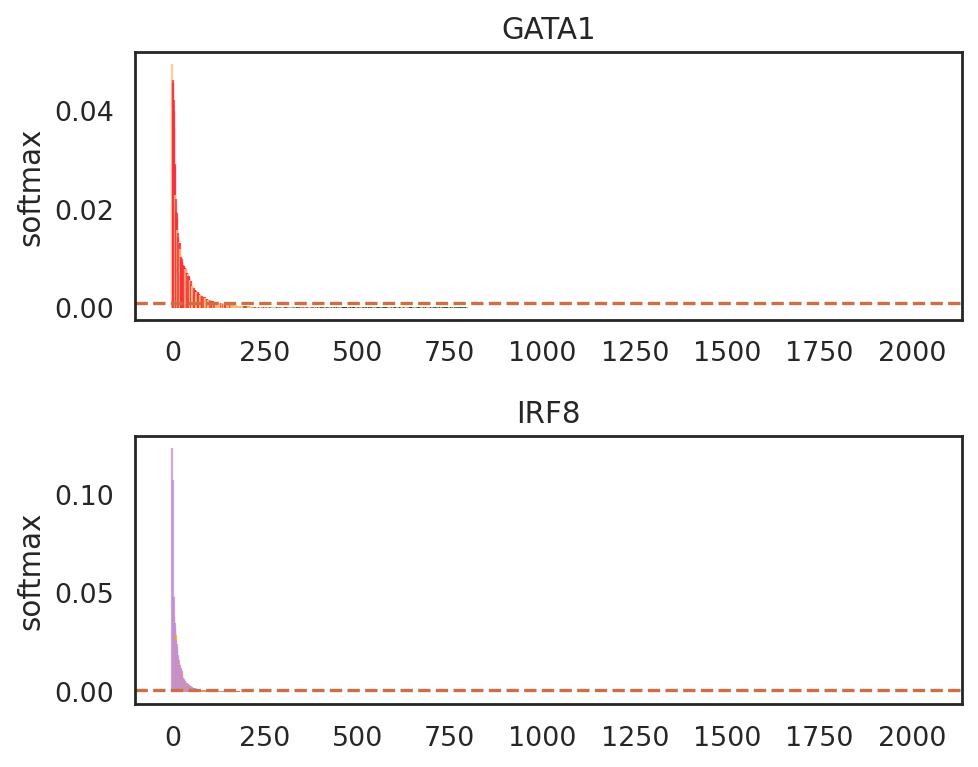
si.pl.entity_barcode(adata_cmp_CM,
layer='softmax',
entities=['GATA1', 'IRF8'],
anno_ref='celltype',
show_cutoff=True,
cutoff=0.001,
palette=palette_celltype,
fig_size=(6, 2.5),
save_fig=False)
[60]:
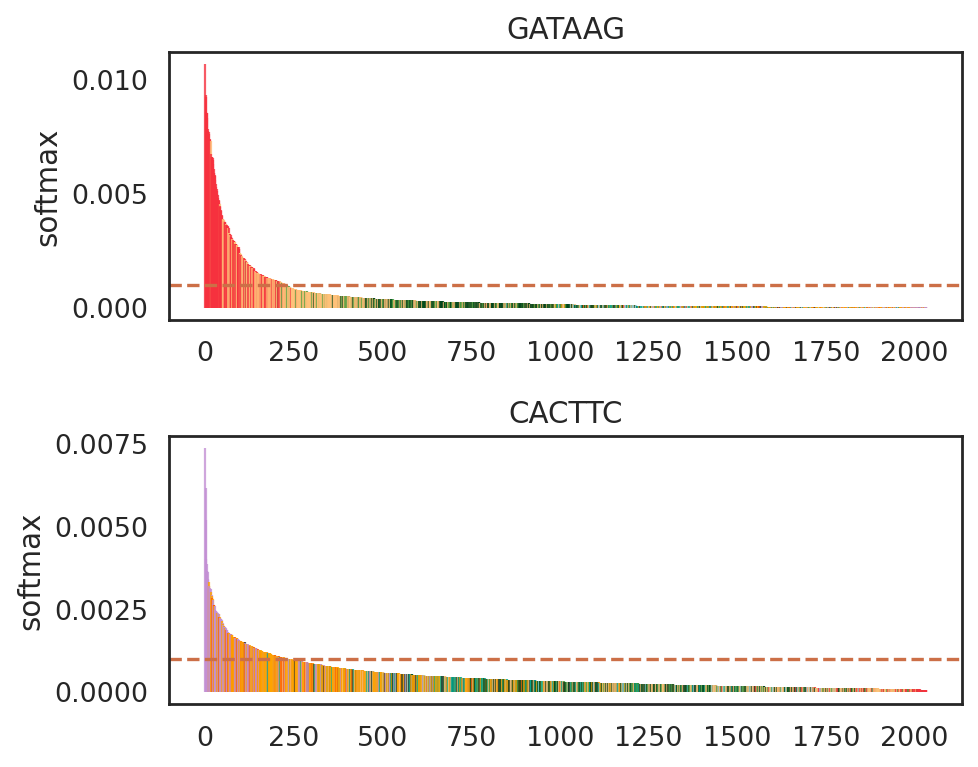
si.pl.entity_barcode(adata_cmp_CK,
layer='softmax',
entities=['GATAAG', 'CACTTC'],
anno_ref='celltype',
show_cutoff=True,
cutoff=0.001,
palette=palette_celltype,
fig_size=(6, 2.5),
save_fig=False)
[ ]:
SIMBA queries
[61]:
si.pl.umap(adata_all[::-1,],
color=['entity_anno'],
dict_palette={'entity_anno': palette_entity_anno},
drawing_order='original',
fig_size=(6.5,5.5))
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
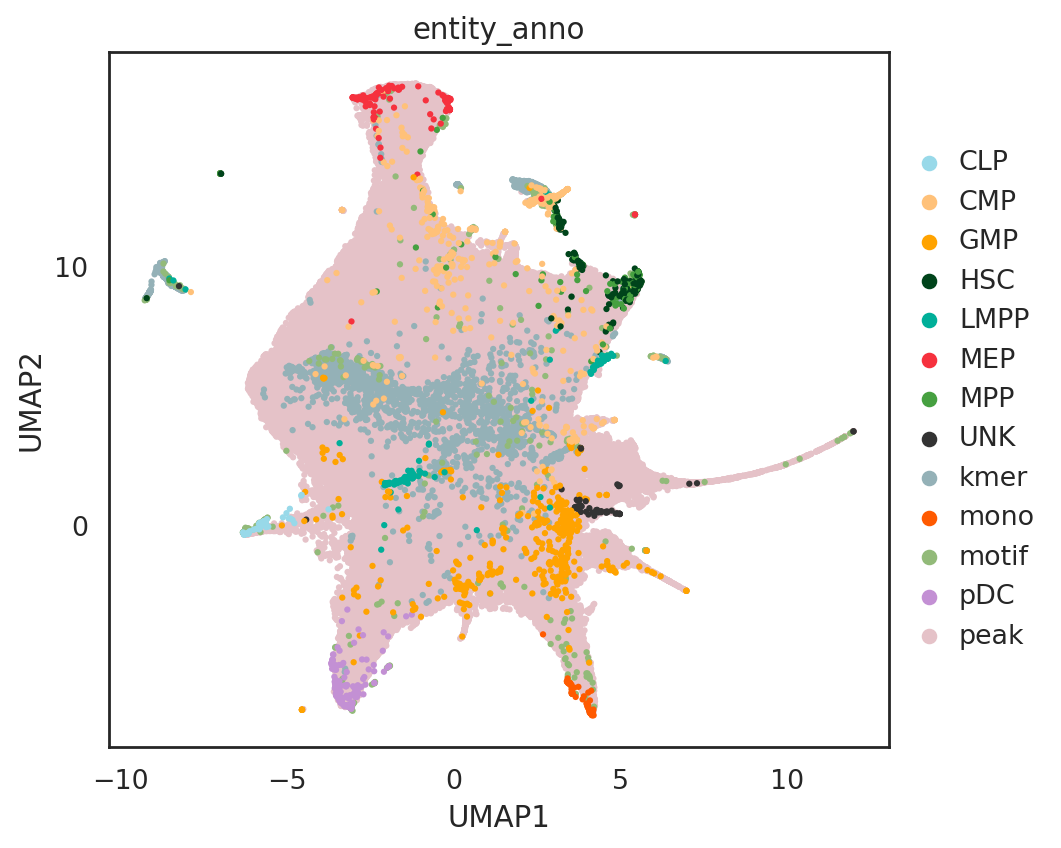
[ ]:
[62]:
# find neighbor motif around the location [-1.5, 16] on UMAP
query_result = si.tl.query(adata_all,
pin=[-1.5,16],
obsm='X_umap',
use_radius=True,r=2,
anno_filter='entity_anno',
filters=['motif', 'kmer'])
print(query_result.shape)
query_result.iloc[:10,]
(31, 5)
[62]:
| celltype | id_dataset | entity_anno | distance | query | |
|---|---|---|---|---|---|
| GATA6 | nan | query_0 | motif | 0.603015 | 0 |
| FOXF2 | nan | query_0 | motif | 0.807087 | 0 |
| GATA1 | nan | query_0 | motif | 0.825012 | 0 |
| GATA2 | nan | query_0 | motif | 0.853963 | 0 |
| GATA4 | nan | query_0 | motif | 0.856798 | 0 |
| GATA3 | nan | query_0 | motif | 0.858775 | 0 |
| GATA1_TAL1 | nan | query_0 | motif | 0.886104 | 0 |
| GATA5 | nan | query_0 | motif | 0.892869 | 0 |
| TTATCA | nan | query_1 | kmer | 0.901073 | 0 |
| GAGATA | nan | query_1 | kmer | 0.904079 | 0 |
[63]:
# show locations of pin point and its neighbor motifs and kmers
si.pl.query(adata_all,
show_texts=False,
color=['entity_anno'], dict_palette={'entity_anno': palette_entity_anno},
alpha=0.9,
alpha_bg=0.1,
fig_size=(6.5,5.5))
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
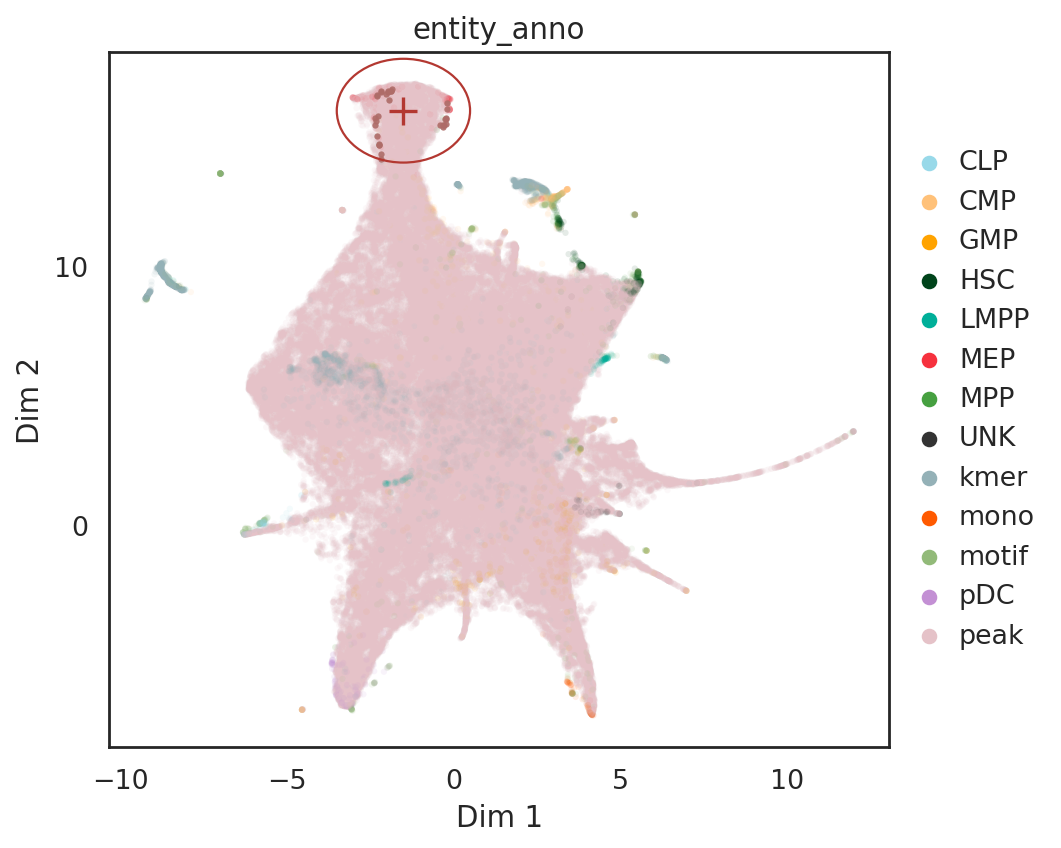
[ ]:
[64]:
# find top 500 neighbor peaks around TF motif 'CEBPB' in SIMBA embedding
query_result = si.tl.query(adata_all,
entity=['CEBPB'],
obsm=None,
use_radius=False,
k=500,
anno_filter='entity_anno',
filters=['peak'])
print(query_result.shape)
query_result.head()
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
(500, 5)
[64]:
| celltype | id_dataset | entity_anno | distance | query | |
|---|---|---|---|---|---|
| chr21_42738544_42739025 | nan | query_2 | peak | 1.004674 | CEBPB |
| chr10_104575728_104576298 | nan | query_2 | peak | 1.016362 | CEBPB |
| chr1_166481044_166481392 | nan | query_2 | peak | 1.030337 | CEBPB |
| chr15_101748029_101748534 | nan | query_2 | peak | 1.046896 | CEBPB |
| chr5_172313932_172314655 | nan | query_2 | peak | 1.047440 | CEBPB |
[65]:
# show locations of neighbor peaks
si.pl.query(adata_all,
show_texts=False,
alpha=0.9,
alpha_bg=0.1,
fig_size=(6.,5.5))
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
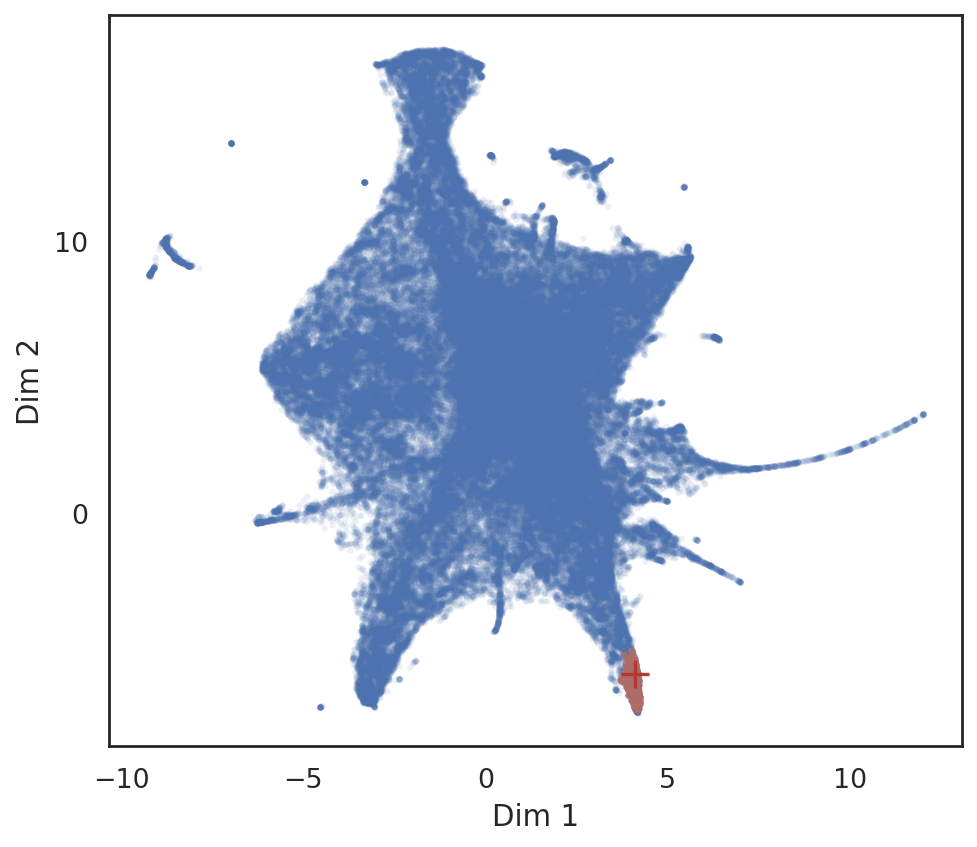
[ ]:
[66]:
# find neighbor entities (both cells and features) of a given TF motif on UMAP
query_result = si.tl.query(adata_all,
entity=['PAX5'],
obsm='X_umap',
use_radius=False,
k=50)
print(query_result.shape)
query_result.iloc[:8,]
(50, 5)
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
[66]:
| celltype | id_dataset | entity_anno | distance | query | |
|---|---|---|---|---|---|
| PAX5 | nan | query_0 | motif | 0.000000 | PAX5 |
| HES2 | nan | query_0 | motif | 0.068228 | PAX5 |
| singles-BM0828-CLP-frozen-151103-20 | CLP | ref | CLP | 0.100645 | PAX5 |
| BM1077-CLP-Frozen-160106-70 | CLP | ref | CLP | 0.117510 | PAX5 |
| BM1077-CLP-Frozen-160106-79 | CLP | ref | CLP | 0.118386 | PAX5 |
| GLIS3 | nan | query_0 | motif | 0.121647 | PAX5 |
| HEY2 | nan | query_0 | motif | 0.122658 | PAX5 |
| HES6 | nan | query_0 | motif | 0.124835 | PAX5 |
[67]:
si.pl.query(adata_all,
obsm='X_umap',
color=['entity_anno'], dict_palette={'entity_anno': palette_entity_anno},
show_texts=False,
alpha=0.9,
alpha_bg=0.1,
fig_size=(6.5,5.5))
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
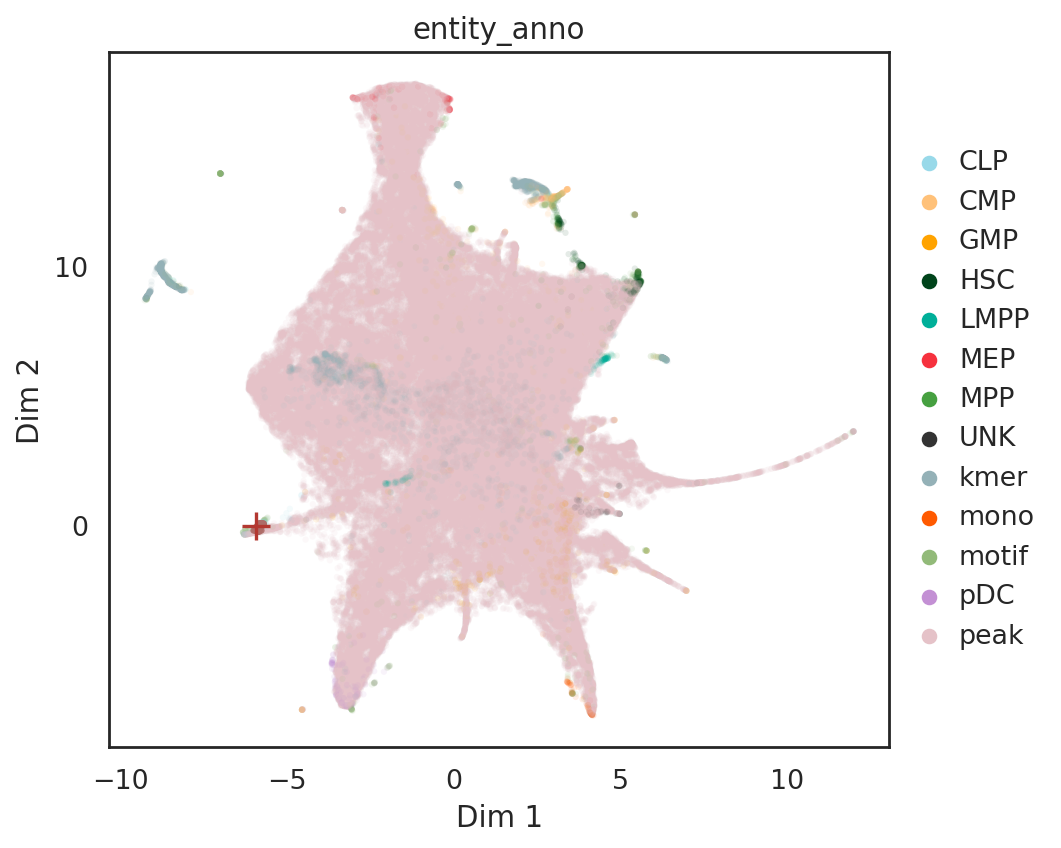
[ ]:
save results
[61]:
adata_CP.write(os.path.join(workdir, 'adata_CP.h5ad'))
adata_PM.write(os.path.join(workdir, 'adata_PM.h5ad'))
adata_PK.write(os.path.join(workdir, 'adata_PK.h5ad'))
adata_C.write(os.path.join(workdir, 'adata_C.h5ad'))
adata_P.write(os.path.join(workdir, 'adata_P.h5ad'))
adata_K.write(os.path.join(workdir, 'adata_K.h5ad'))
adata_M.write(os.path.join(workdir, 'adata_M.h5ad'))
adata_all.write(os.path.join(workdir,'adata_all.h5ad'))
adata_cmp_CM.write(os.path.join(workdir,'adata_cmp_CM.h5ad'))
adata_cmp_CK.write(os.path.join(workdir,'adata_cmp_CK.h5ad'))
... storing 'pbg_id' as categorical
... storing 'pbg_id' as categorical
... storing 'pbg_id' as categorical
... storing 'celltype' as categorical
... storing 'id_dataset' as categorical
... storing 'entity_anno' as categorical
[ ]:
Read back anndata objects
adata_CP = si.read_h5ad(os.path.join(workdir, 'adata_CP.h5ad'))
adata_PM = si.read_h5ad(os.path.join(workdir, 'adata_PM.h5ad'))
adata_PK = si.read_h5ad(os.path.join(workdir, 'adata_PK.h5ad'))
adata_C = si.read_h5ad(os.path.join(workdir, 'adata_C.h5ad'))
adata_P = si.read_h5ad(os.path.join(workdir, 'adata_P.h5ad'))
adata_K = si.read_h5ad(os.path.join(workdir, 'adata_K.h5ad'))
adata_M = si.read_h5ad(os.path.join(workdir, 'adata_M.h5ad'))
adata_all = si.read_h5ad(os.path.join(workdir,'adata_all.h5ad'))
adata_cmp_CM = si.read_h5ad(os.path.join(workdir,'adata_cmp_CM.h5ad'))
adata_cmp_CK = si.read_h5ad(os.path.join(workdir,'adata_cmp_CK.h5ad'))
[ ]: