Batch correction (multiple batches)
Here we will use five scRNA-seq human pancreas datasets of different studies as an example to illustrate how SIMBA performs scRNA-seq batch correction for multiple batches
[1]:
import os
import simba as si
si.__version__
[1]:
'1.0'
[2]:
workdir = 'result_human_pancreas'
si.settings.set_workdir(workdir)
Saving results in: result_human_pancreas
[3]:
si.settings.set_figure_params(dpi=80,
style='white',
fig_size=[5,5],
rc={'image.cmap': 'viridis'})
[4]:
# to make plots prettier
from IPython.display import set_matplotlib_formats
set_matplotlib_formats('retina')
[ ]:
load example data
[5]:
adata_CG_baron = si.datasets.rna_baron2016()
rna_baron2016.h5ad: 0.00B [00:00, ?B/s]
Downloading data ...
rna_baron2016.h5ad: 125MB [01:16, 1.65MB/s]
Downloaded to result_human_pancreas/data.
[6]:
adata_CG_segerstolpe = si.datasets.rna_segerstolpe2016()
rna_segerstolpe2016.h5ad: 0.00B [00:00, ?B/s]
Downloading data ...
rna_segerstolpe2016.h5ad: 98.1MB [01:00, 1.64MB/s]
Downloaded to result_human_pancreas/data.
[7]:
adata_CG_muraro = si.datasets.rna_muraro2016()
rna_muraro2016.h5ad: 0.00B [00:00, ?B/s]
Downloading data ...
rna_muraro2016.h5ad: 82.0MB [00:54, 1.50MB/s]
Downloaded to result_human_pancreas/data.
[8]:
adata_CG_wang = si.datasets.rna_wang2016()
rna_wang2016.h5ad: 0.00B [00:00, ?B/s]
Downloading data ...
rna_wang2016.h5ad: 58.2MB [00:37, 1.57MB/s]
Downloaded to result_human_pancreas/data.
[9]:
adata_CG_xin = si.datasets.rna_xin2016()
rna_xin2016.h5ad: 0.00B [00:00, ?B/s]
Downloading data ...
rna_xin2016.h5ad: 51.3MB [00:33, 1.55MB/s]
Downloaded to result_human_pancreas/data.
[10]:
adata_CG_baron
[10]:
AnnData object with n_obs × n_vars = 8569 × 15558
obs: 'cell_type1'
var: 'feature_symbol'
[11]:
adata_CG_segerstolpe
[11]:
AnnData object with n_obs × n_vars = 2127 × 15558
obs: 'cell_type1'
var: 'feature_symbol'
[12]:
adata_CG_muraro
[12]:
AnnData object with n_obs × n_vars = 2122 × 15558
obs: 'cell_type1'
var: 'feature_symbol'
[13]:
adata_CG_wang
[13]:
AnnData object with n_obs × n_vars = 457 × 15558
obs: 'cell_type1'
var: 'feature_symbol'
[14]:
adata_CG_xin
[14]:
AnnData object with n_obs × n_vars = 1492 × 15558
obs: 'cell_type1'
var: 'feature_symbol'
[ ]:
pre-graph-construction for data baron
[15]:
si.pp.filter_genes(adata_CG_baron,min_n_cells=3)
Before filtering:
8569 cells, 15558 genes
Filter genes based on min_n_cells
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
After filtering out low-expressed genes:
8569 cells, 14689 genes
[16]:
si.pp.cal_qc_rna(adata_CG_baron)
[17]:
si.pl.violin(adata_CG_baron,list_obs=['n_counts','n_genes','pct_mt'])
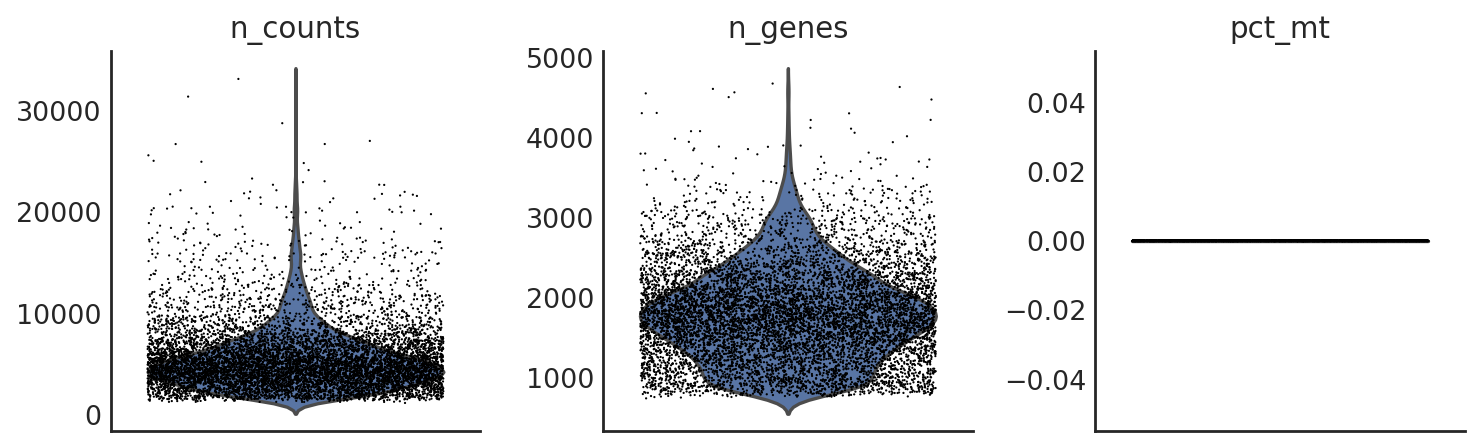
[18]:
si.pp.normalize(adata_CG_baron,method='lib_size')
[19]:
si.pp.log_transform(adata_CG_baron)
[20]:
si.pp.select_variable_genes(adata_CG_baron, n_top_genes=3000)
3000 variable genes are selected.
[21]:
si.pl.variable_genes(adata_CG_baron,show_texts=True,)
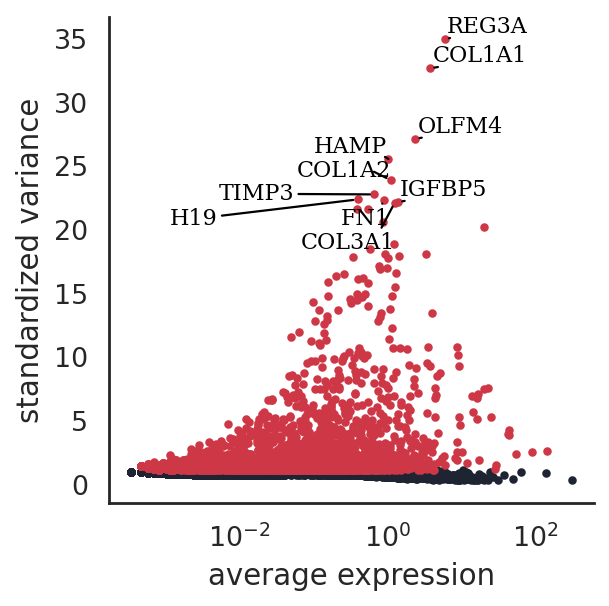
[22]:
si.tl.discretize(adata_CG_baron,n_bins=5)
si.pl.discretize(adata_CG_baron,kde=False)
[0.2638636 1.0367706 1.5327055 2.2109878 3.211969 8.739059 ]
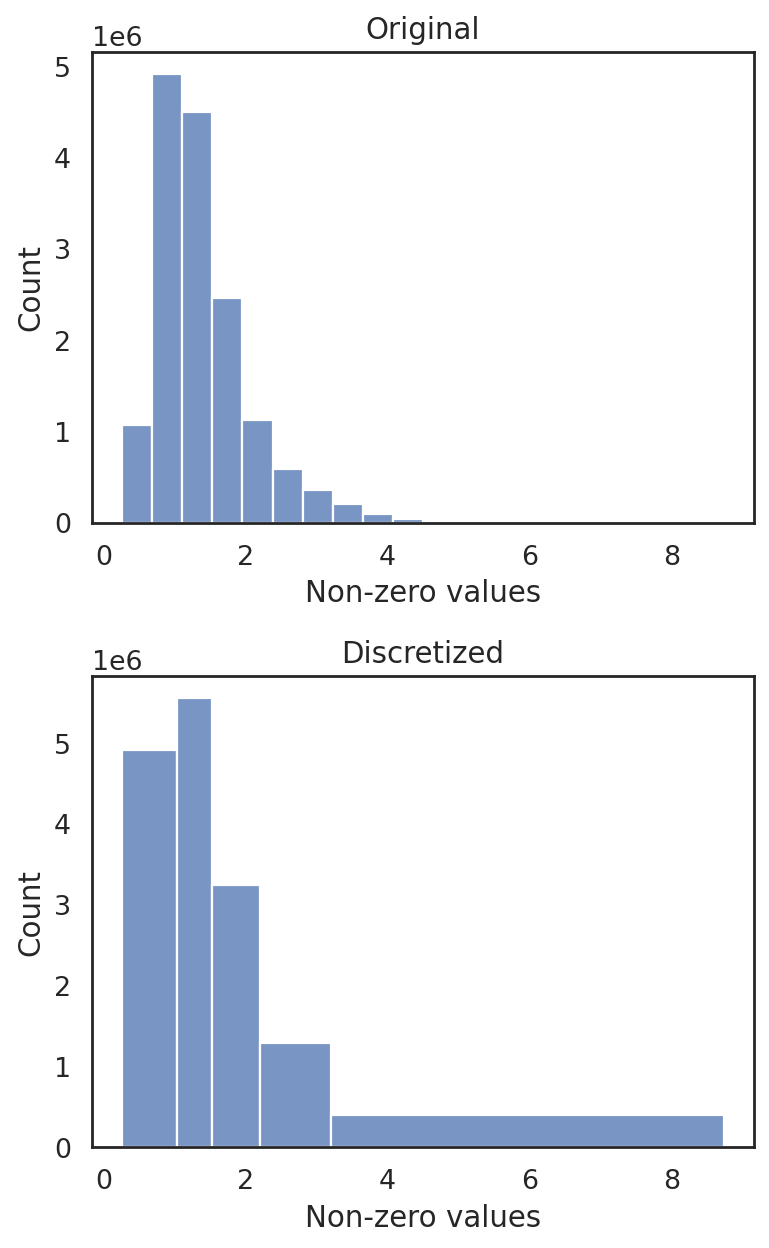
[ ]:
pre-graph-construction for data segerstolpe
[23]:
si.pp.filter_genes(adata_CG_segerstolpe,min_n_cells=3)
Before filtering:
2127 cells, 15558 genes
Filter genes based on min_n_cells
After filtering out low-expressed genes:
2127 cells, 15208 genes
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
[24]:
si.pp.cal_qc_rna(adata_CG_segerstolpe)
[25]:
si.pl.violin(adata_CG_segerstolpe,list_obs=['n_counts','n_genes','pct_mt'])
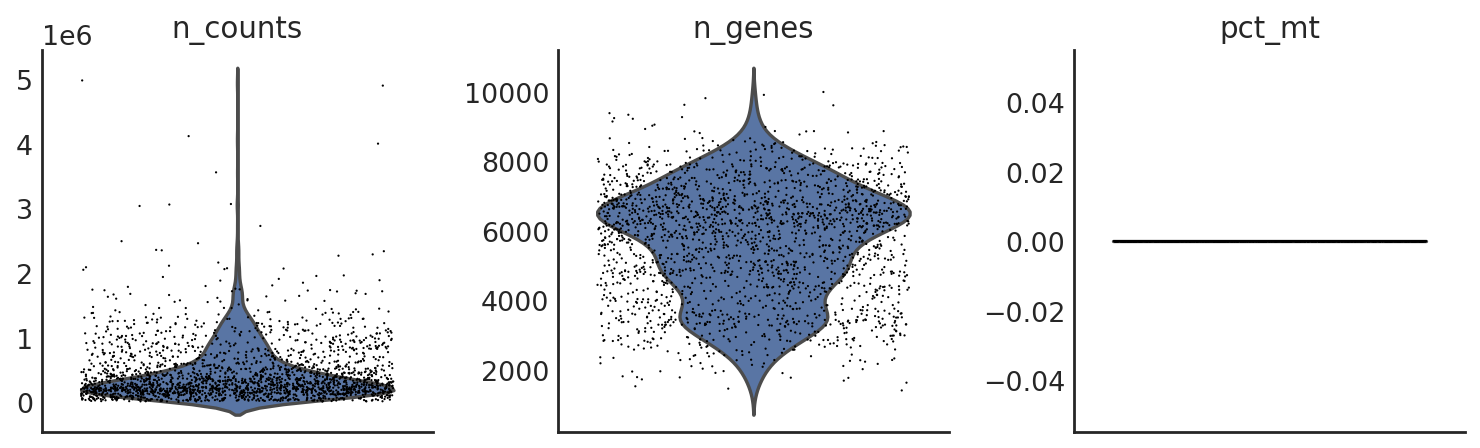
[26]:
si.pp.normalize(adata_CG_segerstolpe,method='lib_size')
[27]:
si.pp.log_transform(adata_CG_segerstolpe)
[28]:
si.pp.select_variable_genes(adata_CG_segerstolpe, n_top_genes=3000)
3000 variable genes are selected.
[29]:
si.pl.variable_genes(adata_CG_segerstolpe,show_texts=True,)
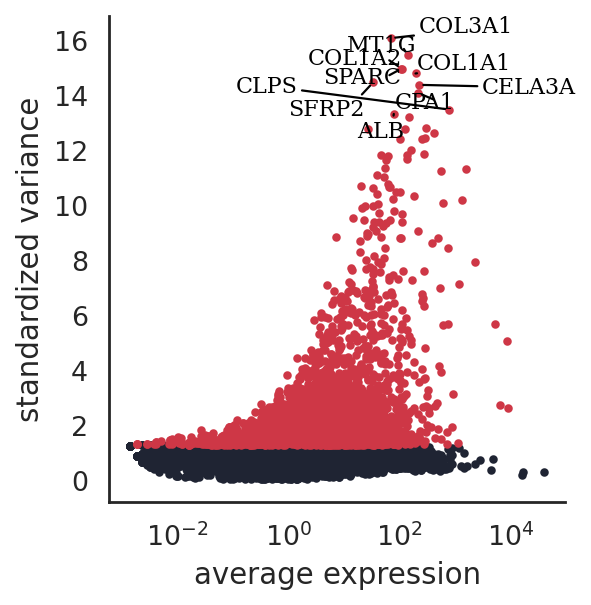
[30]:
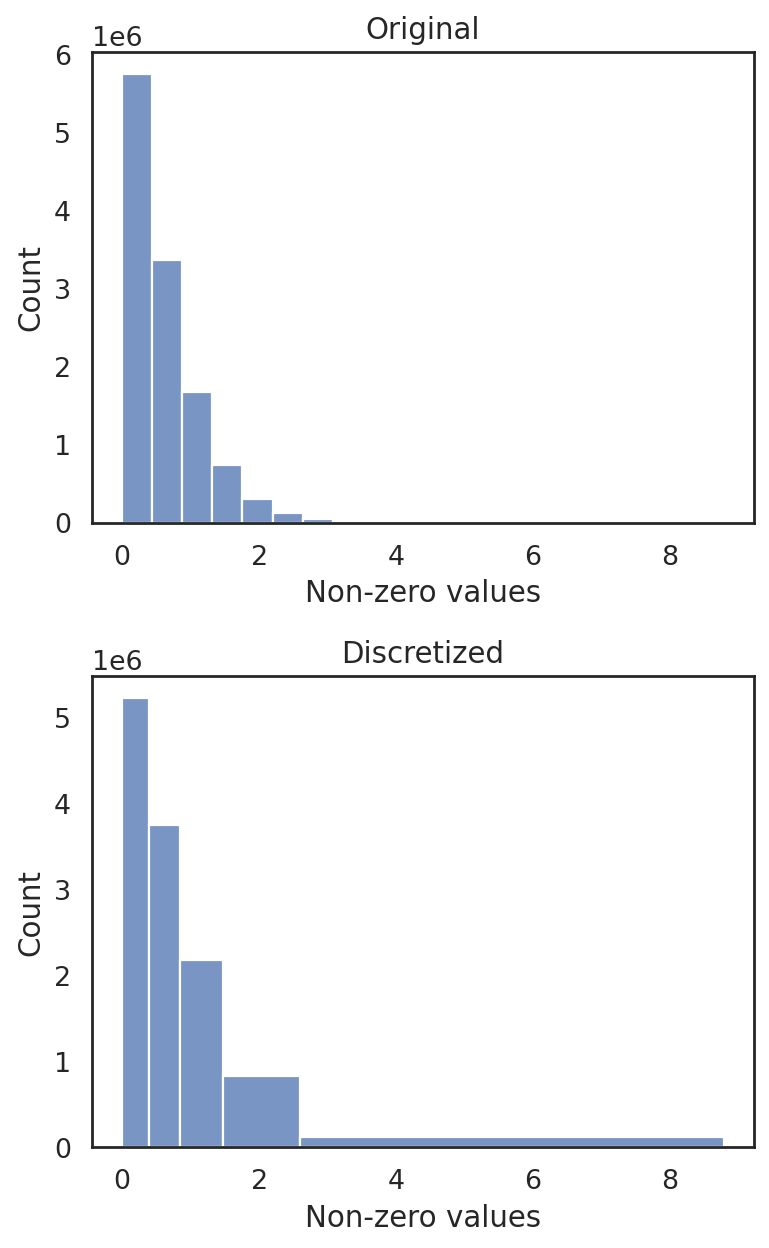
si.tl.discretize(adata_CG_segerstolpe,n_bins=5)
si.pl.discretize(adata_CG_segerstolpe,kde=False)
[2.0057529e-03 3.8856718e-01 8.5258603e-01 1.4747014e+00 2.5910647e+00
8.7814217e+00]
[ ]:
pre-graph-construction for data muraro
[31]:
si.pp.filter_genes(adata_CG_muraro,min_n_cells=3)
Before filtering:
2122 cells, 15558 genes
Filter genes based on min_n_cells
After filtering out low-expressed genes:
2122 cells, 14766 genes
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
[32]:
si.pp.cal_qc_rna(adata_CG_muraro)
[33]:
si.pl.violin(adata_CG_muraro,list_obs=['n_counts','n_genes','pct_mt'])
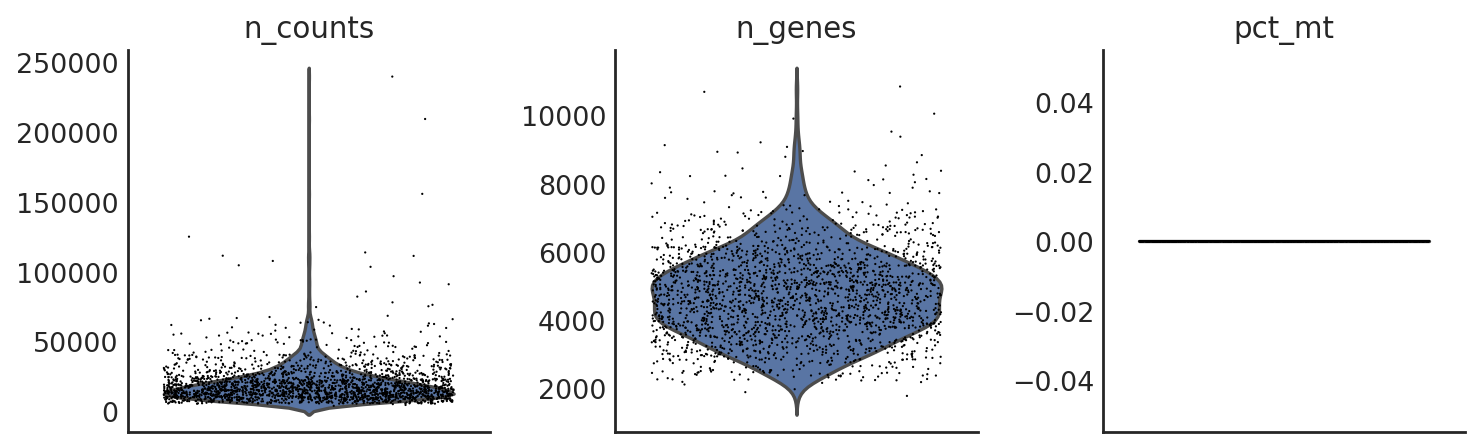
[34]:
si.pp.normalize(adata_CG_muraro,method='lib_size')
[35]:
si.pp.log_transform(adata_CG_muraro)
[36]:
si.pp.select_variable_genes(adata_CG_muraro, n_top_genes=3000)
3000 variable genes are selected.
[37]:
si.pl.variable_genes(adata_CG_muraro,show_texts=True,)
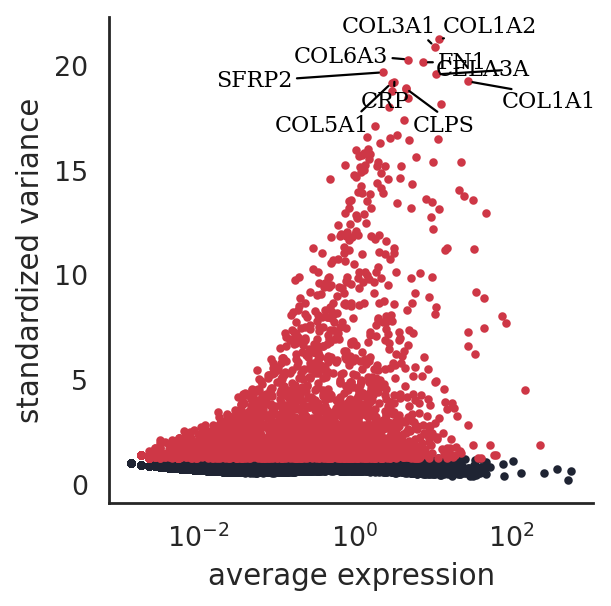
[38]:
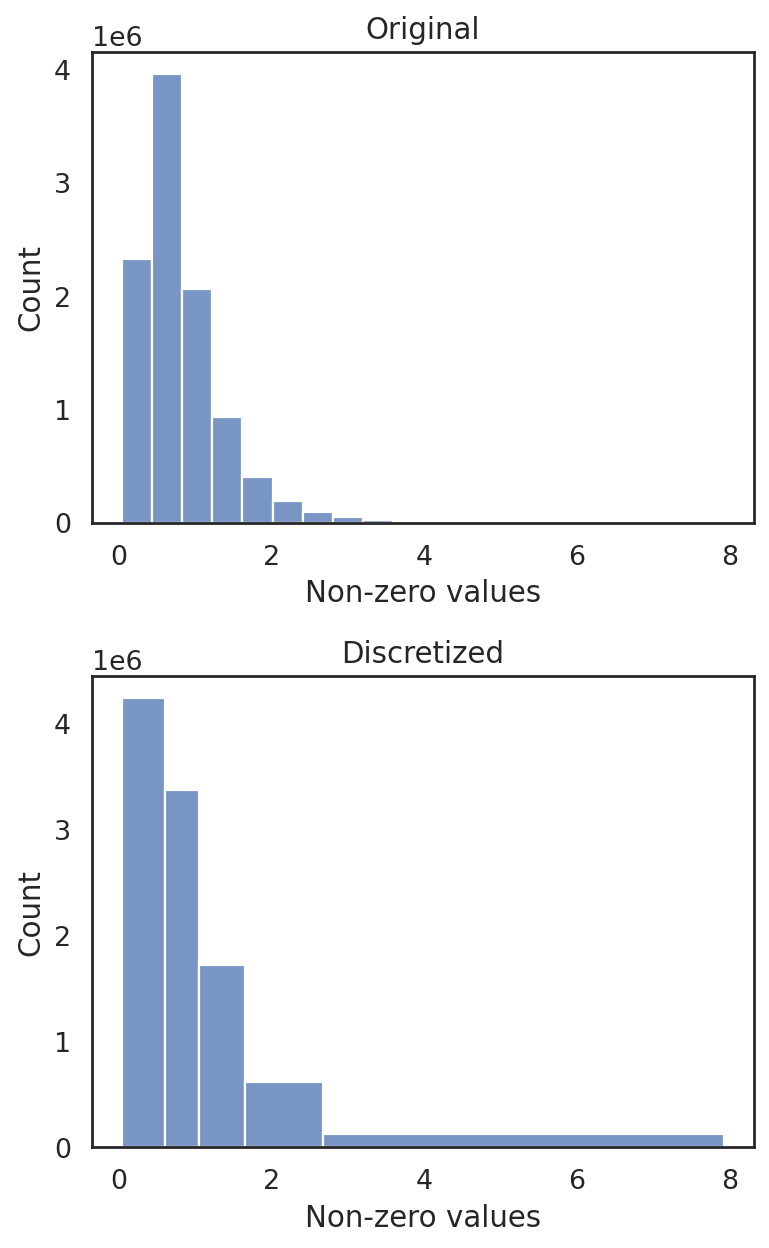
si.tl.discretize(adata_CG_muraro,n_bins=5)
si.pl.discretize(adata_CG_muraro,kde=False)
[0.04096032 0.60292876 1.042481 1.6543355 2.6782749 7.92668 ]
[ ]:
pre-graph-construction for data wang
[39]:
si.pp.filter_genes(adata_CG_wang,min_n_cells=3)
Before filtering:
457 cells, 15558 genes
Filter genes based on min_n_cells
After filtering out low-expressed genes:
457 cells, 14424 genes
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
[40]:
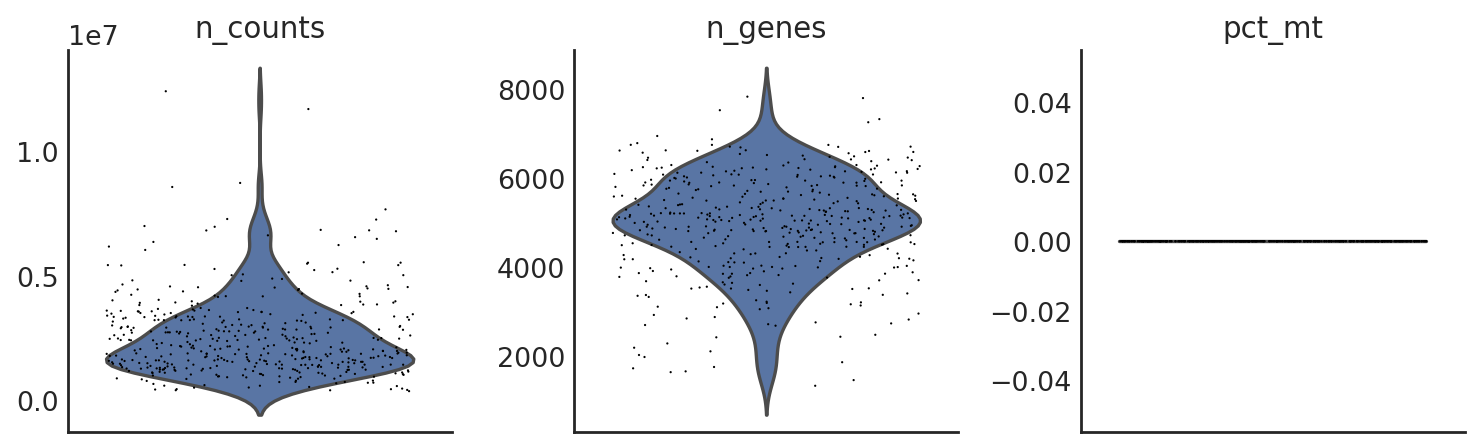
si.pp.cal_qc_rna(adata_CG_wang)
[41]:
si.pl.violin(adata_CG_wang,list_obs=['n_counts','n_genes','pct_mt'])
[42]:
si.pp.normalize(adata_CG_wang,method='lib_size')
[43]:
si.pp.log_transform(adata_CG_wang)
[44]:
si.pp.select_variable_genes(adata_CG_wang, n_top_genes=3000)
3000 variable genes are selected.
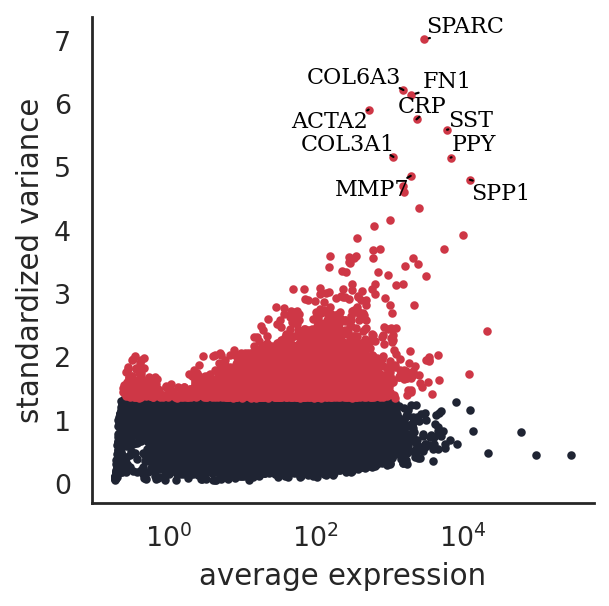
[45]:
si.pl.variable_genes(adata_CG_wang,show_texts=True,)
[46]:
si.tl.discretize(adata_CG_wang,n_bins=5)
si.pl.discretize(adata_CG_wang,kde=False)
[1.4719809e-04 2.6585901e-01 8.4558272e-01 1.5926900e+00 2.6436315e+00
9.0345411e+00]
[ ]:
pre-graph-construction for data xin
[47]:
si.pp.filter_genes(adata_CG_xin,min_n_cells=3)
Before filtering:
1492 cells, 15558 genes
Filter genes based on min_n_cells
After filtering out low-expressed genes:
1492 cells, 13831 genes
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
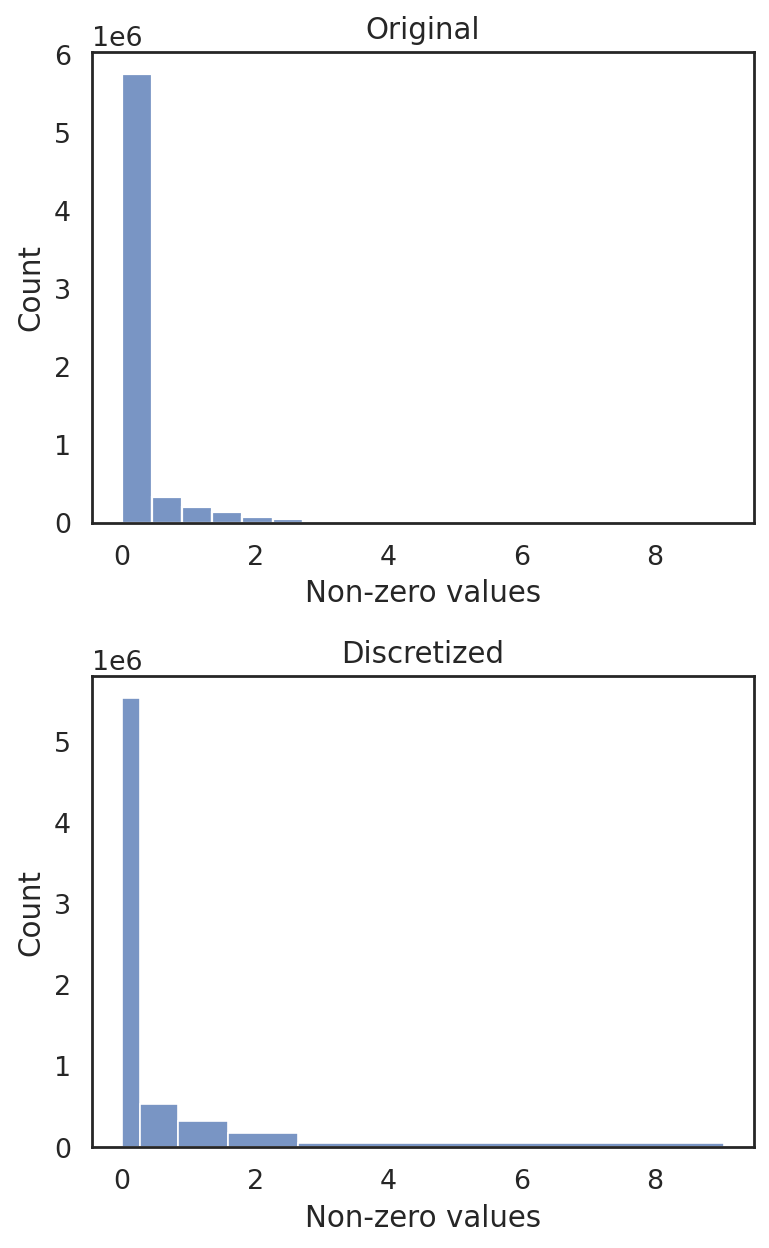
[48]:
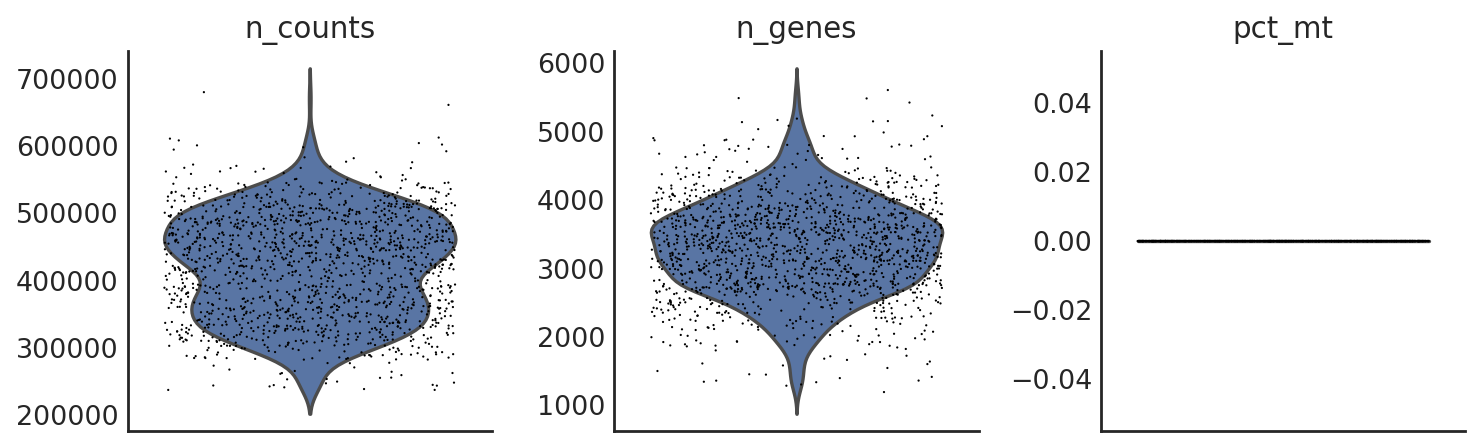
si.pp.cal_qc_rna(adata_CG_xin)
[49]:
si.pl.violin(adata_CG_xin,list_obs=['n_counts','n_genes','pct_mt'])
[50]:
si.pp.normalize(adata_CG_xin,method='lib_size')
[51]:
si.pp.log_transform(adata_CG_xin)
[52]:
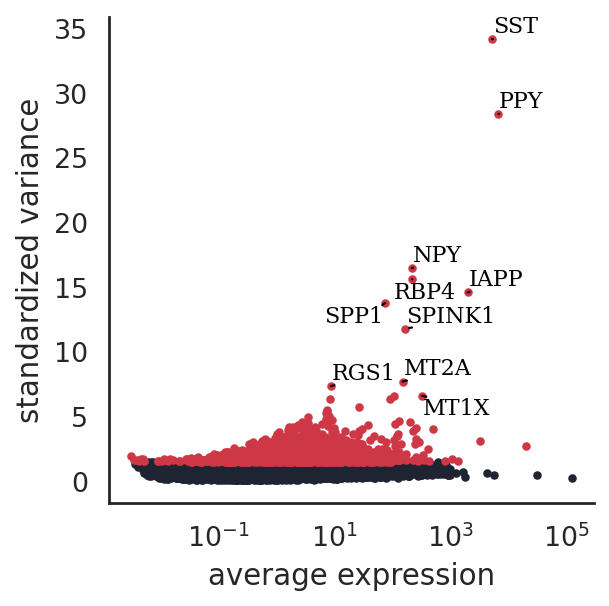
si.pp.select_variable_genes(adata_CG_xin, n_top_genes=3000)
3000 variable genes are selected.
[53]:
si.pl.variable_genes(adata_CG_xin,show_texts=True,)
[54]:
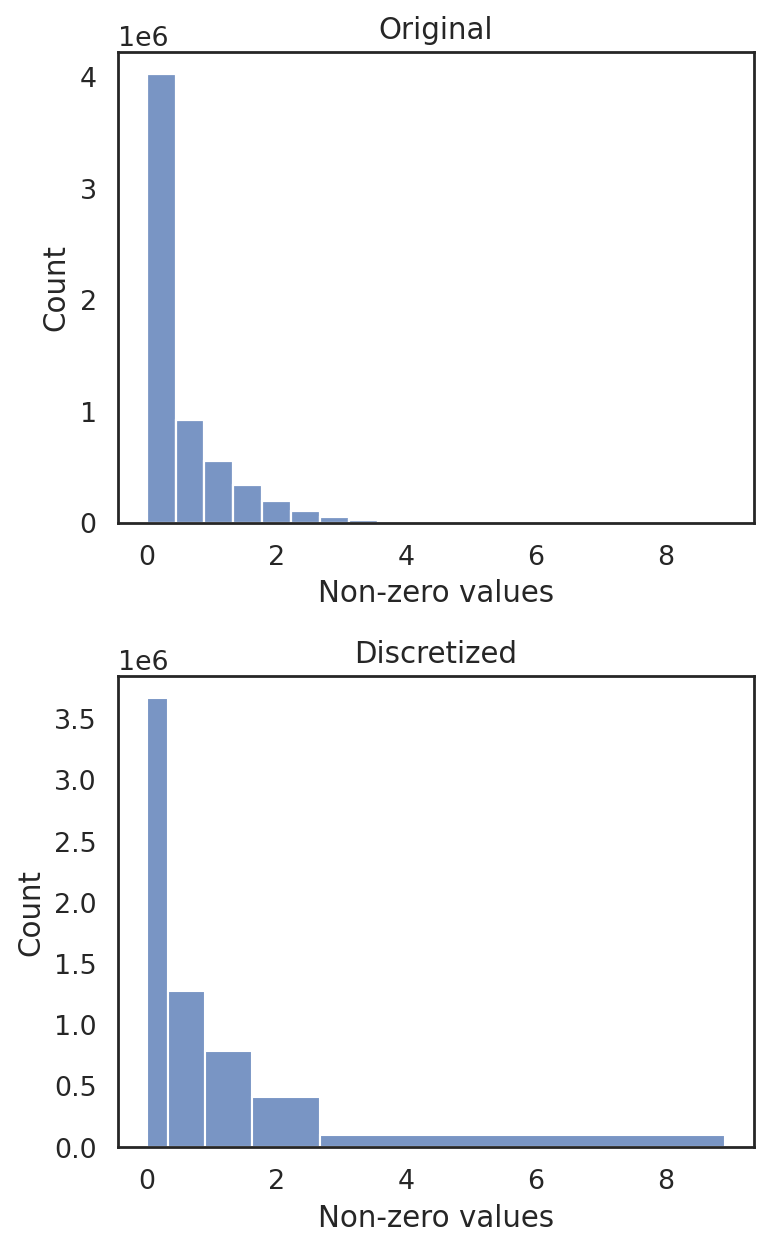
si.tl.discretize(adata_CG_xin,n_bins=5)
si.pl.discretize(adata_CG_xin,kde=False)
[8.2742363e-06 3.3285791e-01 8.9686590e-01 1.6214957e+00 2.6698451e+00
8.9205980e+00]
[ ]:
infer edges between cells of different studies
Edge inference can be performed between any two datasets. Here for simplicity, we always compare each with the largest data (data baron)
[55]:
adata_CbaronCsegerstolpe = si.tl.infer_edges(adata_CG_baron, adata_CG_segerstolpe, n_components=15, k=15)
#shared features: 2966
Performing randomized SVD ...
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
Searching for mutual nearest neighbors ...
25924 edges are selected
[56]:
adata_CbaronCmuraro = si.tl.infer_edges(adata_CG_baron, adata_CG_muraro, n_components=15, k=15)
#shared features: 2894
Performing randomized SVD ...
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
Searching for mutual nearest neighbors ...
26156 edges are selected
[57]:
adata_CbaronCwang = si.tl.infer_edges(adata_CG_baron, adata_CG_wang, n_components=15, k=15)
#shared features: 2707
Performing randomized SVD ...
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
Searching for mutual nearest neighbors ...
6796 edges are selected
[58]:
adata_CbaronCxin = si.tl.infer_edges(adata_CG_baron, adata_CG_xin, n_components=15, k=15)
#shared features: 2405
Performing randomized SVD ...
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
Searching for mutual nearest neighbors ...
20356 edges are selected
[ ]:
generate graph
[59]:
si.tl.gen_graph(list_CG=[adata_CG_baron,
adata_CG_segerstolpe,
adata_CG_muraro,
adata_CG_wang,
adata_CG_xin],
list_CC=[adata_CbaronCsegerstolpe,
adata_CbaronCmuraro,
adata_CbaronCwang,
adata_CbaronCxin
],
copy=False,
dirname='graph0')
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
relation0: source: C, destination: G
#edges: 633924
relation1: source: C, destination: G
#edges: 782354
relation2: source: C, destination: G
#edges: 583741
relation3: source: C, destination: G
#edges: 289621
relation4: source: C, destination: G
#edges: 108049
relation5: source: C2, destination: G
#edges: 572251
relation6: source: C2, destination: G
#edges: 385093
relation7: source: C2, destination: G
#edges: 259491
relation8: source: C2, destination: G
#edges: 143621
relation9: source: C2, destination: G
#edges: 41051
relation10: source: C3, destination: G
#edges: 531261
relation11: source: C3, destination: G
#edges: 429313
relation12: source: C3, destination: G
#edges: 258546
relation13: source: C3, destination: G
#edges: 121782
relation14: source: C3, destination: G
#edges: 38544
relation15: source: C4, destination: G
#edges: 1174840
relation16: source: C4, destination: G
#edges: 89487
relation17: source: C4, destination: G
#edges: 57906
relation18: source: C4, destination: G
#edges: 35627
relation19: source: C4, destination: G
#edges: 13140
relation20: source: C5, destination: G
#edges: 439476
relation21: source: C5, destination: G
#edges: 160233
relation22: source: C5, destination: G
#edges: 118777
relation23: source: C5, destination: G
#edges: 65752
relation24: source: C5, destination: G
#edges: 15367
relation25: source: C, destination: C2
#edges: 25924
relation26: source: C, destination: C3
#edges: 26156
relation27: source: C, destination: C4
#edges: 6796
relation28: source: C, destination: C5
#edges: 20356
Total number of edges: 7428479
Writing graph file "pbg_graph.txt" to "result_human_pancreas/pbg/graph0" ...
Finished.
[ ]:
PBG training
Before PBG training, let’s take a look at the parameters:
[60]:
si.settings.pbg_params
[60]:
{'entity_path': 'result_human_pancreas/pbg/graph0/input/entity',
'edge_paths': ['result_human_pancreas/pbg/graph0/input/edge'],
'checkpoint_path': '',
'entities': {'C': {'num_partitions': 1},
'C2': {'num_partitions': 1},
'C3': {'num_partitions': 1},
'C4': {'num_partitions': 1},
'C5': {'num_partitions': 1},
'G': {'num_partitions': 1}},
'relations': [{'name': 'r0',
'lhs': 'C',
'rhs': 'G',
'operator': 'none',
'weight': 1.0},
{'name': 'r1', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 2.0},
{'name': 'r2', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 3.0},
{'name': 'r3', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 4.0},
{'name': 'r4', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 5.0},
{'name': 'r5', 'lhs': 'C2', 'rhs': 'G', 'operator': 'none', 'weight': 1.0},
{'name': 'r6', 'lhs': 'C2', 'rhs': 'G', 'operator': 'none', 'weight': 2.0},
{'name': 'r7', 'lhs': 'C2', 'rhs': 'G', 'operator': 'none', 'weight': 3.0},
{'name': 'r8', 'lhs': 'C2', 'rhs': 'G', 'operator': 'none', 'weight': 4.0},
{'name': 'r9', 'lhs': 'C2', 'rhs': 'G', 'operator': 'none', 'weight': 5.0},
{'name': 'r10', 'lhs': 'C3', 'rhs': 'G', 'operator': 'none', 'weight': 1.0},
{'name': 'r11', 'lhs': 'C3', 'rhs': 'G', 'operator': 'none', 'weight': 2.0},
{'name': 'r12', 'lhs': 'C3', 'rhs': 'G', 'operator': 'none', 'weight': 3.0},
{'name': 'r13', 'lhs': 'C3', 'rhs': 'G', 'operator': 'none', 'weight': 4.0},
{'name': 'r14', 'lhs': 'C3', 'rhs': 'G', 'operator': 'none', 'weight': 5.0},
{'name': 'r15', 'lhs': 'C4', 'rhs': 'G', 'operator': 'none', 'weight': 1.0},
{'name': 'r16', 'lhs': 'C4', 'rhs': 'G', 'operator': 'none', 'weight': 2.0},
{'name': 'r17', 'lhs': 'C4', 'rhs': 'G', 'operator': 'none', 'weight': 3.0},
{'name': 'r18', 'lhs': 'C4', 'rhs': 'G', 'operator': 'none', 'weight': 4.0},
{'name': 'r19', 'lhs': 'C4', 'rhs': 'G', 'operator': 'none', 'weight': 5.0},
{'name': 'r20', 'lhs': 'C5', 'rhs': 'G', 'operator': 'none', 'weight': 1.0},
{'name': 'r21', 'lhs': 'C5', 'rhs': 'G', 'operator': 'none', 'weight': 2.0},
{'name': 'r22', 'lhs': 'C5', 'rhs': 'G', 'operator': 'none', 'weight': 3.0},
{'name': 'r23', 'lhs': 'C5', 'rhs': 'G', 'operator': 'none', 'weight': 4.0},
{'name': 'r24', 'lhs': 'C5', 'rhs': 'G', 'operator': 'none', 'weight': 5.0},
{'name': 'r25', 'lhs': 'C', 'rhs': 'C2', 'operator': 'none', 'weight': 10.0},
{'name': 'r26', 'lhs': 'C', 'rhs': 'C3', 'operator': 'none', 'weight': 10.0},
{'name': 'r27', 'lhs': 'C', 'rhs': 'C4', 'operator': 'none', 'weight': 10.0},
{'name': 'r28',
'lhs': 'C',
'rhs': 'C5',
'operator': 'none',
'weight': 10.0}],
'dynamic_relations': False,
'dimension': 50,
'global_emb': False,
'comparator': 'dot',
'num_epochs': 10,
'workers': 4,
'num_batch_negs': 50,
'num_uniform_negs': 50,
'loss_fn': 'softmax',
'lr': 0.1,
'early_stopping': False,
'regularization_coef': 0.0,
'wd': 0.0,
'wd_interval': 50,
'eval_fraction': 0.05,
'eval_num_batch_negs': 50,
'eval_num_uniform_negs': 50,
'checkpoint_preservation_interval': None}
[ ]:
If no parameters need to be adjusted, the training can be simply done with:
si.tl.pbg_train(auto_wd=True, save_wd=True, output='model')
[ ]:
Here we show how to adjust training-related parameters if needed. In general, weight decay wd is the only parameter that might need to be adjusted based on the following pbg metric plots. However, in almost all the cases, the automatically decided wd (enabling it by setting auto_wd=True) works well.
E.g. we want to change the number of cpus workers:
[61]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
# dict_config['wd'] = 0.00477
dict_config['workers'] = 12
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model')
Auto-estimated weight decay is 0.00477
`.settings.pbg_params['wd']` has been updated to 0.00477
Converting input data ...
[2021-06-29 21:40:01.574447] Using the 29 relation types given in the config
[2021-06-29 21:40:01.574932] Searching for the entities in the edge files...
[2021-06-29 21:40:11.294029] Entity type C:
[2021-06-29 21:40:11.295167] - Found 8569 entities
[2021-06-29 21:40:11.295475] - Removing the ones with fewer than 1 occurrences...
[2021-06-29 21:40:11.296746] - Left with 8569 entities
[2021-06-29 21:40:11.297024] - Shuffling them...
[2021-06-29 21:40:11.302851] Entity type C2:
[2021-06-29 21:40:11.303206] - Found 2127 entities
[2021-06-29 21:40:11.303454] - Removing the ones with fewer than 1 occurrences...
[2021-06-29 21:40:11.303926] - Left with 2127 entities
[2021-06-29 21:40:11.304210] - Shuffling them...
[2021-06-29 21:40:11.305721] Entity type C3:
[2021-06-29 21:40:11.306009] - Found 2122 entities
[2021-06-29 21:40:11.306265] - Removing the ones with fewer than 1 occurrences...
[2021-06-29 21:40:11.306737] - Left with 2122 entities
[2021-06-29 21:40:11.306998] - Shuffling them...
[2021-06-29 21:40:11.308515] Entity type C4:
[2021-06-29 21:40:11.308790] - Found 457 entities
[2021-06-29 21:40:11.309949] - Removing the ones with fewer than 1 occurrences...
[2021-06-29 21:40:11.310271] - Left with 457 entities
[2021-06-29 21:40:11.310552] - Shuffling them...
[2021-06-29 21:40:11.311062] Entity type C5:
[2021-06-29 21:40:11.311327] - Found 1492 entities
[2021-06-29 21:40:11.311585] - Removing the ones with fewer than 1 occurrences...
[2021-06-29 21:40:11.311987] - Left with 1492 entities
[2021-06-29 21:40:11.312252] - Shuffling them...
[2021-06-29 21:40:11.313398] Entity type G:
[2021-06-29 21:40:11.313660] - Found 7988 entities
[2021-06-29 21:40:11.313927] - Removing the ones with fewer than 1 occurrences...
[2021-06-29 21:40:11.315054] - Left with 7988 entities
[2021-06-29 21:40:11.315325] - Shuffling them...
[2021-06-29 21:40:11.320490] Preparing counts and dictionaries for entities and relation types:
[2021-06-29 21:40:11.329341] - Writing count of entity type C and partition 0
[2021-06-29 21:40:11.347993] - Writing count of entity type C2 and partition 0
[2021-06-29 21:40:11.358701] - Writing count of entity type C3 and partition 0
[2021-06-29 21:40:11.372474] - Writing count of entity type C4 and partition 0
[2021-06-29 21:40:11.383678] - Writing count of entity type C5 and partition 0
[2021-06-29 21:40:11.394194] - Writing count of entity type G and partition 0
[2021-06-29 21:40:11.419719] Preparing edge path result_human_pancreas/pbg/graph0/input/edge, out of the edges found in result_human_pancreas/pbg/graph0/pbg_graph.txt
using fast version
[2021-06-29 21:40:11.422970] Taking the fast train!
[2021-06-29 21:40:11.925852] - Processed 100000 edges so far...
[2021-06-29 21:40:12.412556] - Processed 200000 edges so far...
[2021-06-29 21:40:12.900183] - Processed 300000 edges so far...
[2021-06-29 21:40:13.404772] - Processed 400000 edges so far...
[2021-06-29 21:40:13.890525] - Processed 500000 edges so far...
[2021-06-29 21:40:14.378665] - Processed 600000 edges so far...
[2021-06-29 21:40:14.868879] - Processed 700000 edges so far...
[2021-06-29 21:40:15.356932] - Processed 800000 edges so far...
[2021-06-29 21:40:15.853686] - Processed 900000 edges so far...
[2021-06-29 21:40:16.343524] - Processed 1000000 edges so far...
[2021-06-29 21:40:16.832229] - Processed 1100000 edges so far...
[2021-06-29 21:40:17.323025] - Processed 1200000 edges so far...
[2021-06-29 21:40:17.810497] - Processed 1300000 edges so far...
[2021-06-29 21:40:18.300955] - Processed 1400000 edges so far...
[2021-06-29 21:40:18.788931] - Processed 1500000 edges so far...
[2021-06-29 21:40:19.276166] - Processed 1600000 edges so far...
[2021-06-29 21:40:19.759103] - Processed 1700000 edges so far...
[2021-06-29 21:40:20.240909] - Processed 1800000 edges so far...
[2021-06-29 21:40:20.724873] - Processed 1900000 edges so far...
[2021-06-29 21:40:21.212335] - Processed 2000000 edges so far...
[2021-06-29 21:40:21.696513] - Processed 2100000 edges so far...
[2021-06-29 21:40:22.181341] - Processed 2200000 edges so far...
[2021-06-29 21:40:22.666027] - Processed 2300000 edges so far...
[2021-06-29 21:40:23.151720] - Processed 2400000 edges so far...
[2021-06-29 21:40:23.636540] - Processed 2500000 edges so far...
[2021-06-29 21:40:24.121809] - Processed 2600000 edges so far...
[2021-06-29 21:40:24.612813] - Processed 2700000 edges so far...
[2021-06-29 21:40:25.109549] - Processed 2800000 edges so far...
[2021-06-29 21:40:25.597482] - Processed 2900000 edges so far...
[2021-06-29 21:40:26.082887] - Processed 3000000 edges so far...
[2021-06-29 21:40:26.568924] - Processed 3100000 edges so far...
[2021-06-29 21:40:27.062947] - Processed 3200000 edges so far...
[2021-06-29 21:40:27.567539] - Processed 3300000 edges so far...
[2021-06-29 21:40:28.054611] - Processed 3400000 edges so far...
[2021-06-29 21:40:28.544680] - Processed 3500000 edges so far...
[2021-06-29 21:40:29.035308] - Processed 3600000 edges so far...
[2021-06-29 21:40:29.524765] - Processed 3700000 edges so far...
[2021-06-29 21:40:30.014224] - Processed 3800000 edges so far...
[2021-06-29 21:40:30.504160] - Processed 3900000 edges so far...
[2021-06-29 21:40:30.991183] - Processed 4000000 edges so far...
[2021-06-29 21:40:31.488787] - Processed 4100000 edges so far...
[2021-06-29 21:40:31.980574] - Processed 4200000 edges so far...
[2021-06-29 21:40:32.472833] - Processed 4300000 edges so far...
[2021-06-29 21:40:32.962001] - Processed 4400000 edges so far...
[2021-06-29 21:40:33.450430] - Processed 4500000 edges so far...
[2021-06-29 21:40:33.937078] - Processed 4600000 edges so far...
[2021-06-29 21:40:34.428206] - Processed 4700000 edges so far...
[2021-06-29 21:40:34.914289] - Processed 4800000 edges so far...
[2021-06-29 21:40:35.398330] - Processed 4900000 edges so far...
[2021-06-29 21:40:35.885293] - Processed 5000000 edges so far...
[2021-06-29 21:40:36.391198] - Processed 5100000 edges so far...
[2021-06-29 21:40:36.878443] - Processed 5200000 edges so far...
[2021-06-29 21:40:37.359751] - Processed 5300000 edges so far...
[2021-06-29 21:40:37.843185] - Processed 5400000 edges so far...
[2021-06-29 21:40:38.327391] - Processed 5500000 edges so far...
[2021-06-29 21:40:38.811798] - Processed 5600000 edges so far...
[2021-06-29 21:40:39.296875] - Processed 5700000 edges so far...
[2021-06-29 21:40:39.785000] - Processed 5800000 edges so far...
[2021-06-29 21:40:40.271171] - Processed 5900000 edges so far...
[2021-06-29 21:40:40.757481] - Processed 6000000 edges so far...
[2021-06-29 21:40:41.242214] - Processed 6100000 edges so far...
[2021-06-29 21:40:41.724551] - Processed 6200000 edges so far...
[2021-06-29 21:40:42.205805] - Processed 6300000 edges so far...
[2021-06-29 21:40:42.689184] - Processed 6400000 edges so far...
[2021-06-29 21:40:43.172650] - Processed 6500000 edges so far...
[2021-06-29 21:40:43.658179] - Processed 6600000 edges so far...
[2021-06-29 21:40:44.145839] - Processed 6700000 edges so far...
[2021-06-29 21:40:44.635070] - Processed 6800000 edges so far...
[2021-06-29 21:40:45.127950] - Processed 6900000 edges so far...
[2021-06-29 21:40:45.615272] - Processed 7000000 edges so far...
[2021-06-29 21:40:46.102117] - Processed 7100000 edges so far...
[2021-06-29 21:40:46.589956] - Processed 7200000 edges so far...
[2021-06-29 21:40:47.076644] - Processed 7300000 edges so far...
[2021-06-29 21:40:47.569635] - Processed 7400000 edges so far...
[2021-06-29 21:40:55.369168] - Processed 7428479 edges in total
Starting training ...
Finished
[ ]:
If
wdis specified by users instead of being automatically decided, then make sure to update it in simba setting:
si.settings.pbg_params = dict_config.copy()
[ ]:
The trained result can be loaded in with the following steps:
By default, it’s using the current training result stored in .setting.pbg_params
# load in graph ('graph0') info
si.load_graph_stats()
# load in model info for ('graph0')
si.load_pbg_config()
Users can also specify different pathss
# load in graph ('graph0') info
si.load_graph_stats(path='./result_human_pancreas/pbg/graph0/')
# load in model info for ('graph0')
si.load_pbg_config(path='./result_human_pancreas/pbg/graph0/model/')
[ ]:
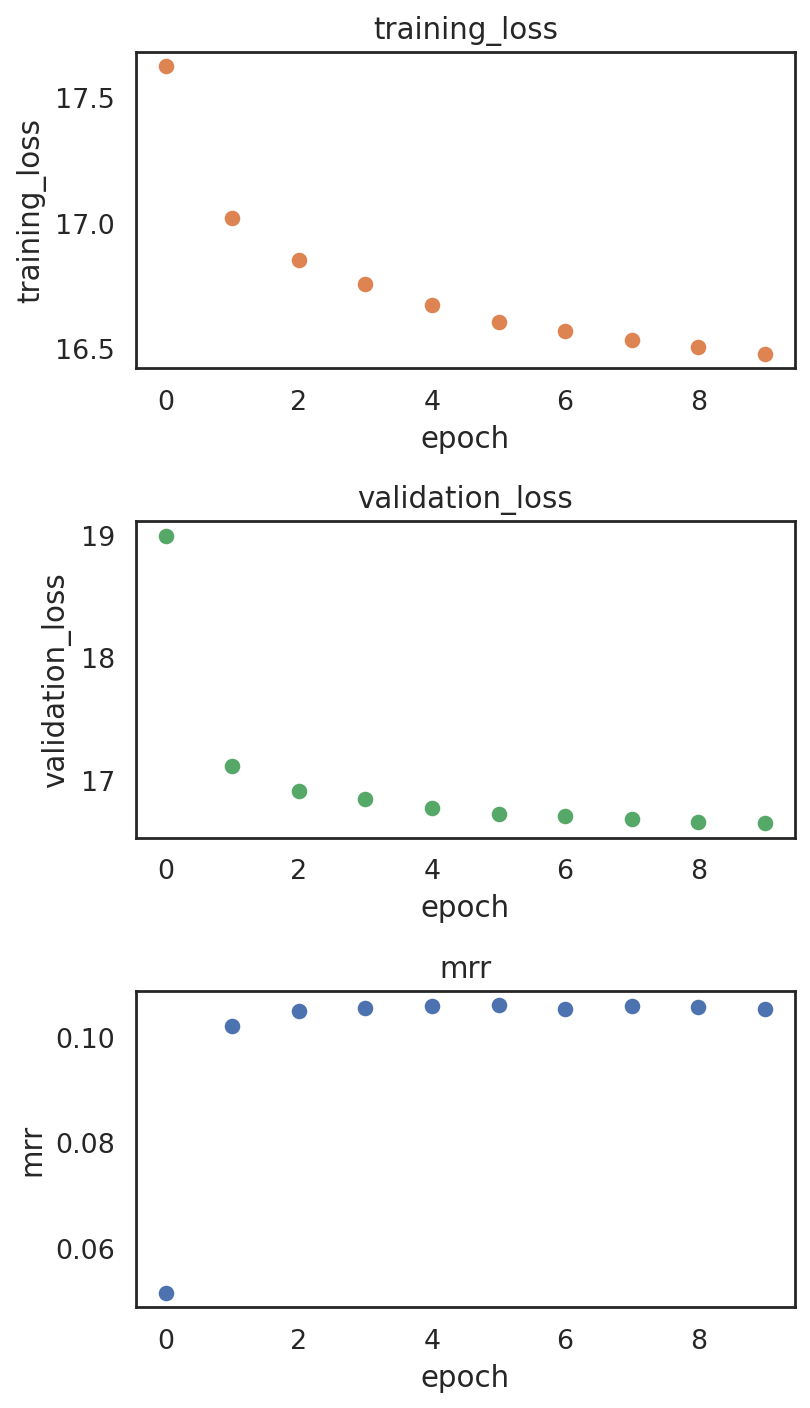
Plotting training metrics to make sure the model is not overfitting
[62]:
si.pl.pbg_metrics(fig_ncol=1)
[ ]:
Post-training Analysis
[63]:
dict_adata = si.read_embedding()
[64]:
dict_adata
[64]:
{'C': AnnData object with n_obs × n_vars = 8569 × 50,
'C2': AnnData object with n_obs × n_vars = 2127 × 50,
'C3': AnnData object with n_obs × n_vars = 2122 × 50,
'C4': AnnData object with n_obs × n_vars = 457 × 50,
'G': AnnData object with n_obs × n_vars = 7988 × 50,
'C5': AnnData object with n_obs × n_vars = 1492 × 50}
[65]:
adata_C = dict_adata['C'] # embeddings for cells from data baron
adata_C2 = dict_adata['C2'] # embeddings for cells from data segerstolpe
adata_C3 = dict_adata['C3'] # embeddings for cells from data muraro
adata_C4 = dict_adata['C4'] # embeddings for cells from data wang
adata_C5 = dict_adata['C5'] # embeddings for cells from data xin
adata_G = dict_adata['G'] # embeddings for genes
[66]:
adata_C
[66]:
AnnData object with n_obs × n_vars = 8569 × 50
[67]:
adata_C2
[67]:
AnnData object with n_obs × n_vars = 2127 × 50
[68]:
adata_C3
[68]:
AnnData object with n_obs × n_vars = 2122 × 50
[69]:
adata_C4
[69]:
AnnData object with n_obs × n_vars = 457 × 50
[70]:
adata_C5
[70]:
AnnData object with n_obs × n_vars = 1492 × 50
[71]:
adata_G
[71]:
AnnData object with n_obs × n_vars = 7988 × 50
[ ]:
visualize embeddings of cells (data baron)
[72]:
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CG_baron[adata_C.obs_names,:].obs['cell_type1'].copy()
adata_C
[72]:
AnnData object with n_obs × n_vars = 8569 × 50
obs: 'celltype'
[73]:
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
[74]:
si.pl.umap(adata_C,
color=['celltype'],
fig_size=(5,4),
drawing_order='random')
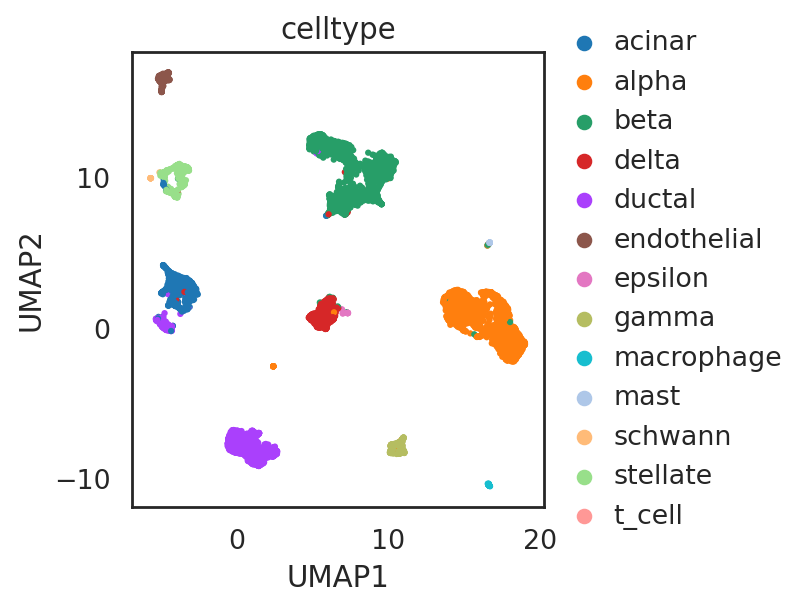
visualize embeddings of cells (data segerstolpe)
[75]:
## Add annotation of celltypes (optional)
adata_C2.obs['celltype'] = adata_CG_segerstolpe[adata_C2.obs_names,:].obs['cell_type1'].copy()
adata_C2
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
[75]:
AnnData object with n_obs × n_vars = 2127 × 50
obs: 'celltype'
[76]:
si.tl.umap(adata_C2,n_neighbors=15,n_components=2)
[77]:
adata_C2
[77]:
AnnData object with n_obs × n_vars = 2127 × 50
obs: 'celltype'
obsm: 'X_umap'
[79]:
si.pl.umap(adata_C2,
color=['celltype'],
fig_size=(5,4),
drawing_order='random')
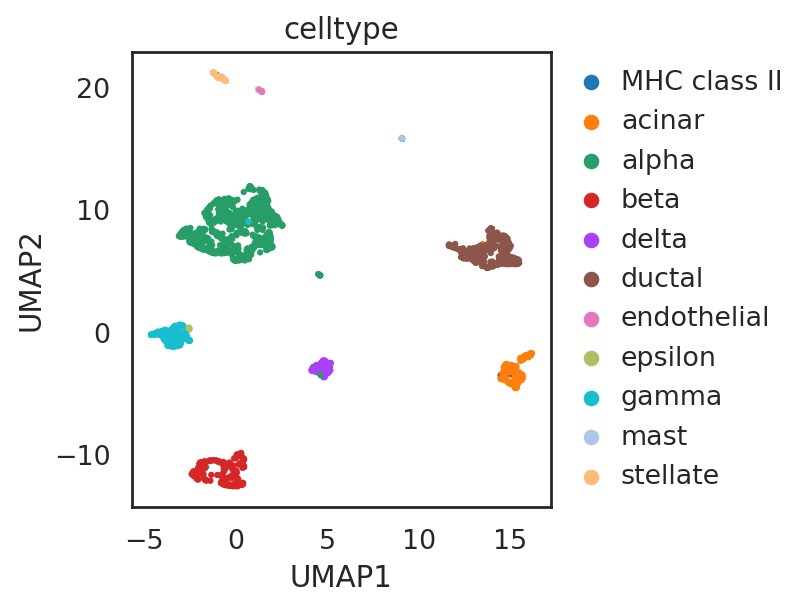
[ ]:
visualize embeddings of cells (data muraro)
[80]:
## Add annotation of celltypes (optional)
adata_C3.obs['celltype'] = adata_CG_muraro[adata_C3.obs_names,:].obs['cell_type1'].copy()
adata_C3
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
[80]:
AnnData object with n_obs × n_vars = 2122 × 50
obs: 'celltype'
[81]:
si.tl.umap(adata_C3,n_neighbors=15,n_components=2)
[82]:
si.pl.umap(adata_C3,
color=['celltype'],
fig_size=(5,4),
drawing_order='random')
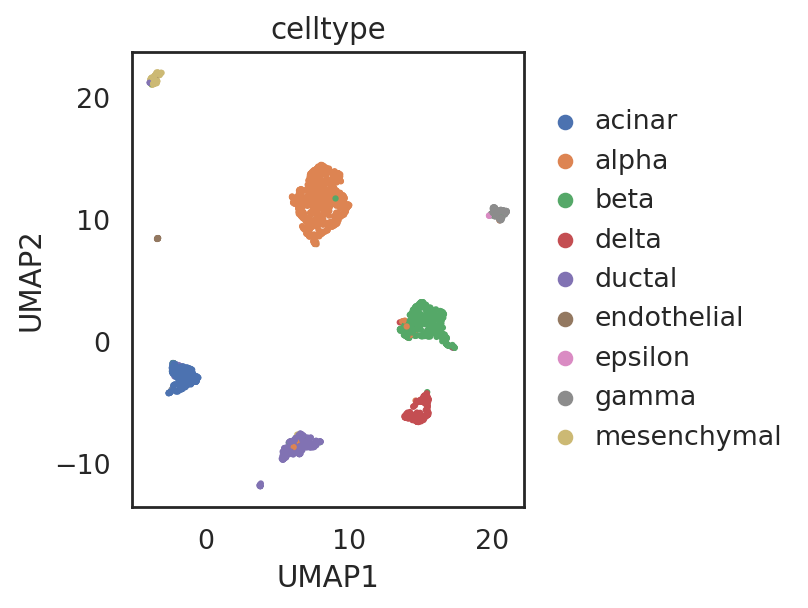
[ ]:
visualize embeddings of cells (data wang)
[83]:
## Add annotation of celltypes (optional)
adata_C4.obs['celltype'] = adata_CG_wang[adata_C4.obs_names,:].obs['cell_type1'].copy()
adata_C4
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
[83]:
AnnData object with n_obs × n_vars = 457 × 50
obs: 'celltype'
[84]:
si.tl.umap(adata_C4,n_neighbors=15,n_components=2)
[85]:
si.pl.umap(adata_C4,
color=['celltype'],
fig_size=(5,4),
drawing_order='random')
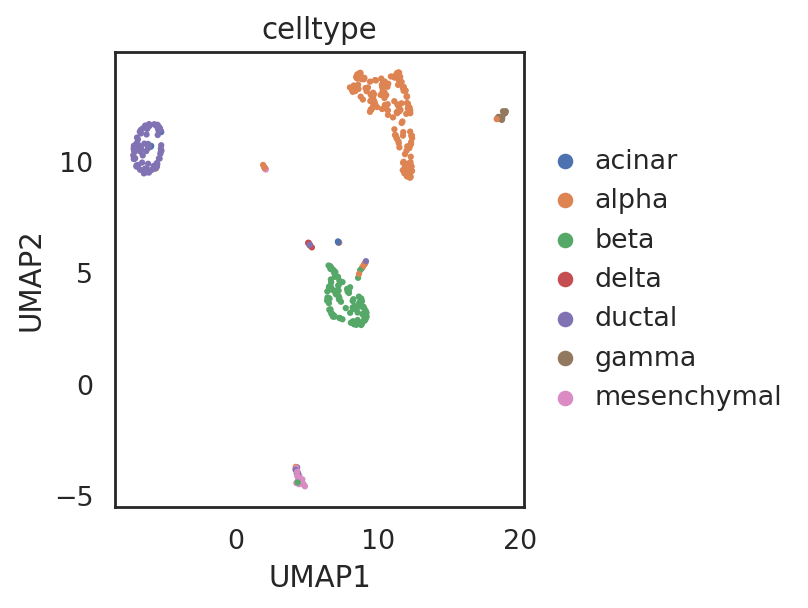
[ ]:
visualize embeddings of cells (data xin)
[86]:
## Add annotation of celltypes (optional)
adata_C5.obs['celltype'] = adata_CG_xin[adata_C5.obs_names,:].obs['cell_type1'].copy()
adata_C5
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
[86]:
AnnData object with n_obs × n_vars = 1492 × 50
obs: 'celltype'
[87]:
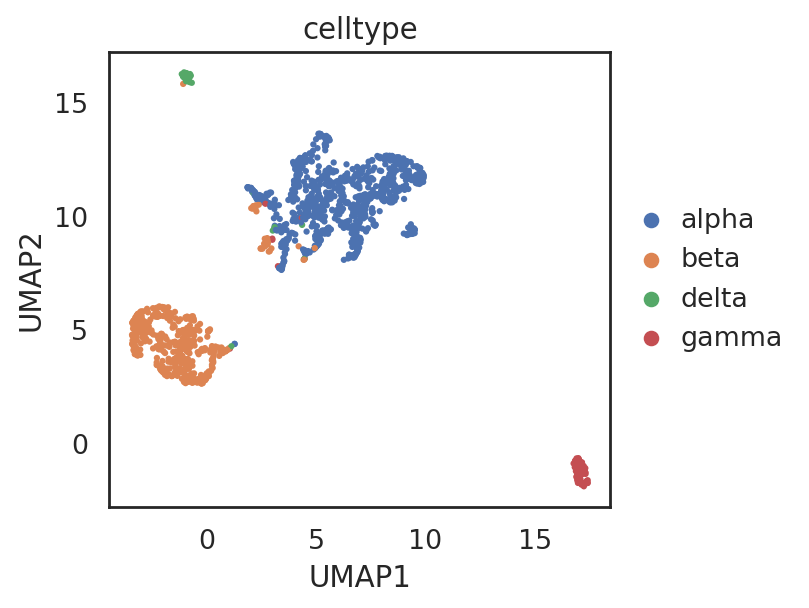
si.tl.umap(adata_C5,n_neighbors=15,n_components=2)
[89]:
si.pl.umap(adata_C5,
color=['celltype'],
fig_size=(5,4),
drawing_order='random')
[ ]:
visualize co-embeddings of all batches
[90]:
# we choose the largest dataset as a reference
adata_all = si.tl.embed(adata_ref=adata_C,
list_adata_query=[adata_C2, adata_C3, adata_C4, adata_C5],
use_precomputed=False)
Performing softmax transformation for query data 0;
Performing softmax transformation for query data 1;
Performing softmax transformation for query data 2;
Performing softmax transformation for query data 3;
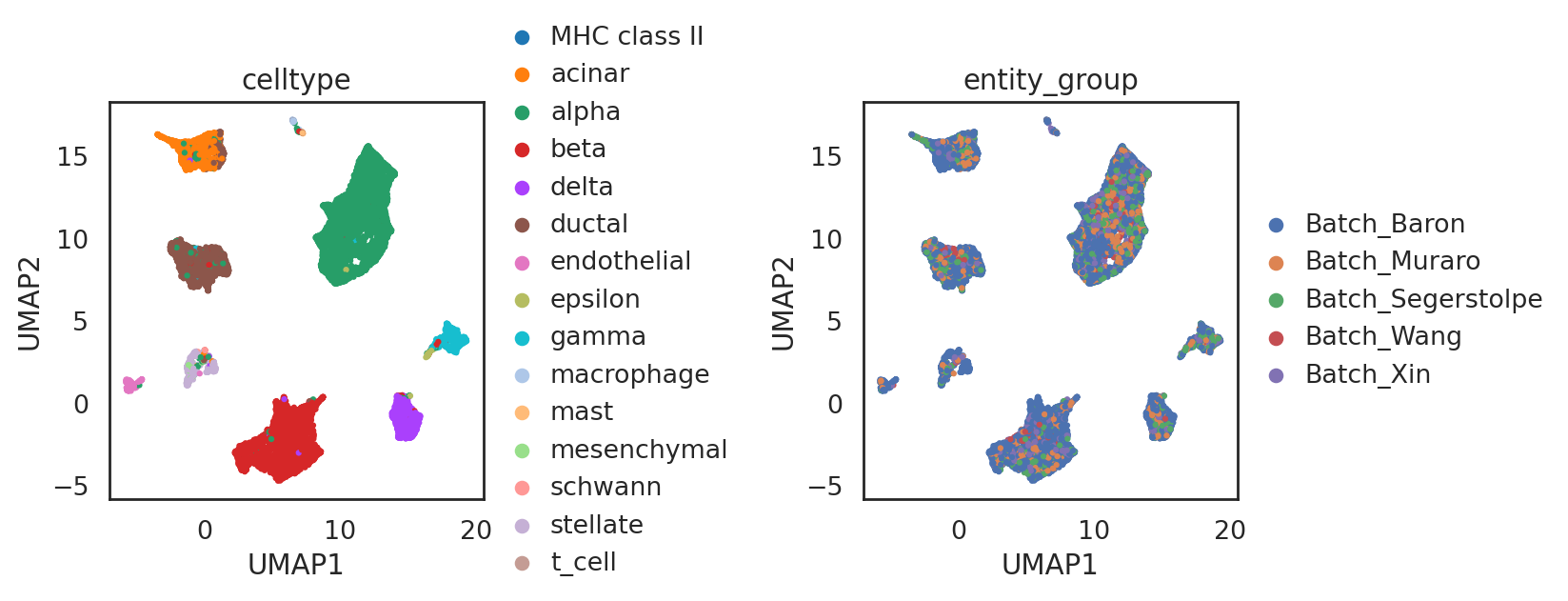
[91]:
## add annotations of two batches
adata_all.obs['entity_group'] = ""
adata_all.obs.loc[adata_C.obs_names, 'entity_group'] = "Batch_Baron"
adata_all.obs.loc[adata_C2.obs_names, 'entity_group'] = "Batch_Segerstolpe"
adata_all.obs.loc[adata_C3.obs_names, 'entity_group'] = "Batch_Muraro"
adata_all.obs.loc[adata_C4.obs_names, 'entity_group'] = "Batch_Wang"
adata_all.obs.loc[adata_C5.obs_names, 'entity_group'] = "Batch_Xin"
adata_all.obs.head()
[91]:
| celltype | id_dataset | entity_group | |
|---|---|---|---|
| human3_lib1.final_cell_0442 | alpha | ref | Batch_Baron |
| human4_lib3.final_cell_0006 | ductal | ref | Batch_Baron |
| human3_lib1.final_cell_0749 | stellate | ref | Batch_Baron |
| human3_lib2.final_cell_0485 | alpha | ref | Batch_Baron |
| human1_lib2.final_cell_0243 | delta | ref | Batch_Baron |
[92]:
si.tl.umap(adata_all,n_neighbors=15,n_components=2)
[94]:
si.pl.umap(adata_all,color=['celltype','entity_group'],
drawing_order='random',
fig_size=(5,4))
[ ]:
visualize embeddings of cells and genes
[95]:
adata_all_CG = si.tl.embed(adata_ref=adata_C,
list_adata_query=[adata_C2, adata_C3, adata_C4, adata_C5, adata_G],
use_precomputed=False)
Performing softmax transformation for query data 0;
Performing softmax transformation for query data 1;
Performing softmax transformation for query data 2;
Performing softmax transformation for query data 3;
Performing softmax transformation for query data 4;
[96]:
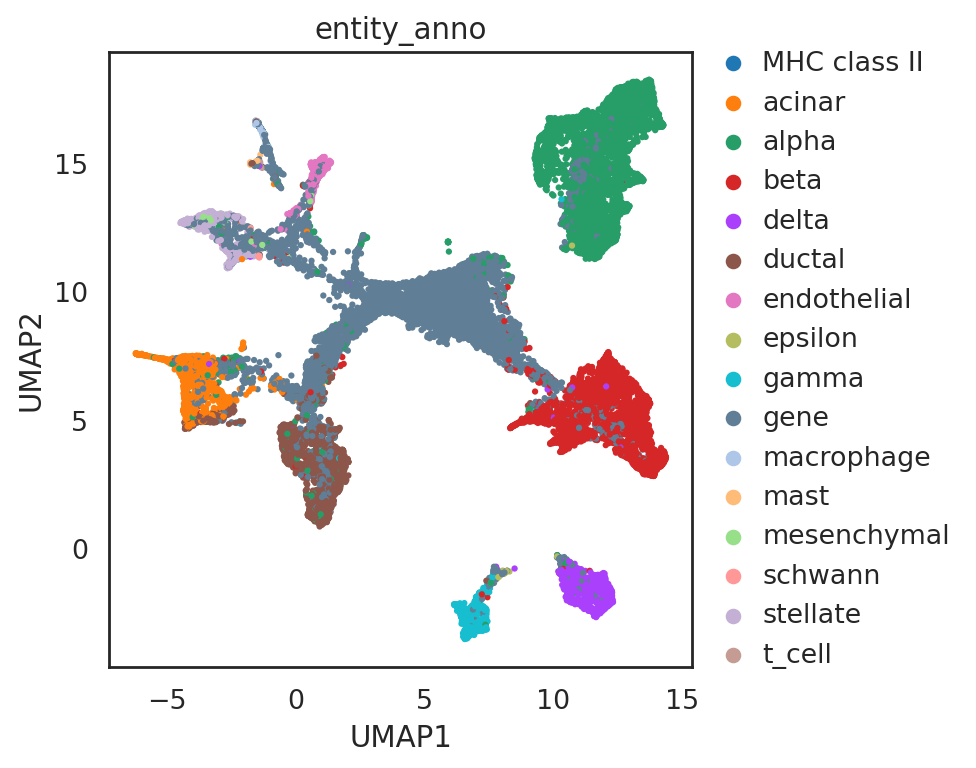
## add annotations of all entities
adata_all_CG.obs['entity_anno'] = ""
adata_all_CG.obs.loc[adata_C.obs_names, 'entity_anno'] = adata_all_CG.obs.loc[adata_C.obs_names, 'celltype'].tolist()
adata_all_CG.obs.loc[adata_C2.obs_names, 'entity_anno'] = adata_all_CG.obs.loc[adata_C2.obs_names, 'celltype'].tolist()
adata_all_CG.obs.loc[adata_C3.obs_names, 'entity_anno'] = adata_all_CG.obs.loc[adata_C3.obs_names, 'celltype'].tolist()
adata_all_CG.obs.loc[adata_C4.obs_names, 'entity_anno'] = adata_all_CG.obs.loc[adata_C4.obs_names, 'celltype'].tolist()
adata_all_CG.obs.loc[adata_C5.obs_names, 'entity_anno'] = adata_all_CG.obs.loc[adata_C5.obs_names, 'celltype'].tolist()
adata_all_CG.obs.loc[adata_G.obs_names, 'entity_anno'] = 'gene'
adata_all_CG.obs.head()
[96]:
| celltype | id_dataset | entity_anno | |
|---|---|---|---|
| human3_lib1.final_cell_0442 | alpha | ref | alpha |
| human4_lib3.final_cell_0006 | ductal | ref | ductal |
| human3_lib1.final_cell_0749 | stellate | ref | stellate |
| human3_lib2.final_cell_0485 | alpha | ref | alpha |
| human1_lib2.final_cell_0243 | delta | ref | delta |
[97]:
palette_celltype = {'MHC class II': '#1f77b4', 'acinar': '#ff7f0e',
'alpha': '#279e68',
'beta': '#d62728',
'delta': '#aa40fc',
'ductal': '#8c564b',
'endothelial': '#e377c2',
'epsilon': '#b5bd61',
'gamma': '#17becf',
'macrophage': '#aec7e8',
'mast': '#ffbb78',
'mesenchymal': '#98df8a',
'schwann': '#ff9896',
'stellate': '#c5b0d5',
't_cell': '#c49c94'}
palette_entity_anno = palette_celltype.copy()
palette_entity_anno['gene'] = "#607e95"
[99]:
si.tl.umap(adata_all_CG,n_neighbors=15,n_components=2)
[100]:
si.pl.umap(adata_all_CG,
color=['entity_anno'],
dict_palette={'entity_anno': palette_entity_anno},
drawing_order='random',
fig_size=(6,5))
[101]:
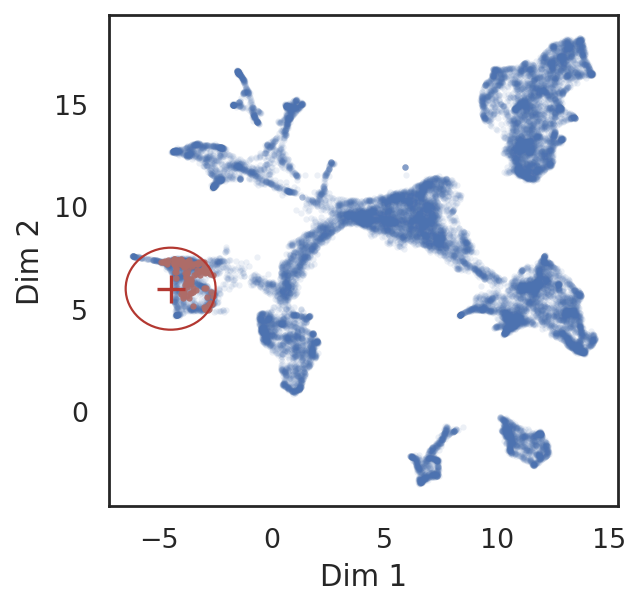
# find neighbor genes of given pins
query_genes = si.tl.query(adata_all_CG,
pin=[-4.5, 6],
use_radius=True, r=2,
obsm='X_umap',
anno_filter='entity_anno',
filters=['gene'])
print(query_genes.shape)
query_genes.head()
(132, 5)
[101]:
| celltype | id_dataset | entity_anno | distance | query | |
|---|---|---|---|---|---|
| FGL1 | NaN | query_4 | gene | 0.516078 | 0 |
| ALB | NaN | query_4 | gene | 0.531156 | 0 |
| ITIH4 | NaN | query_4 | gene | 0.543160 | 0 |
| ALDOB | NaN | query_4 | gene | 0.549413 | 0 |
| PLIN5 | NaN | query_4 | gene | 0.565572 | 0 |
[102]:
# show locations of pin points and its neighbor genes
si.pl.query(adata_all_CG,
show_texts=False,
alpha=0.9,
alpha_bg=0.1,
fig_legend_ncol=1,
fig_size=(4,4))
[103]:
si.pl.umap(adata_all_CG[::-1],
color=['entity_anno'],
dict_palette={'entity_anno': palette_entity_anno},
drawing_order='original',
show_texts=True,
texts=query_genes.index[:10],
text_expand=(1.2,1.4),
fig_size=(6,5),
fig_legend_ncol=1)
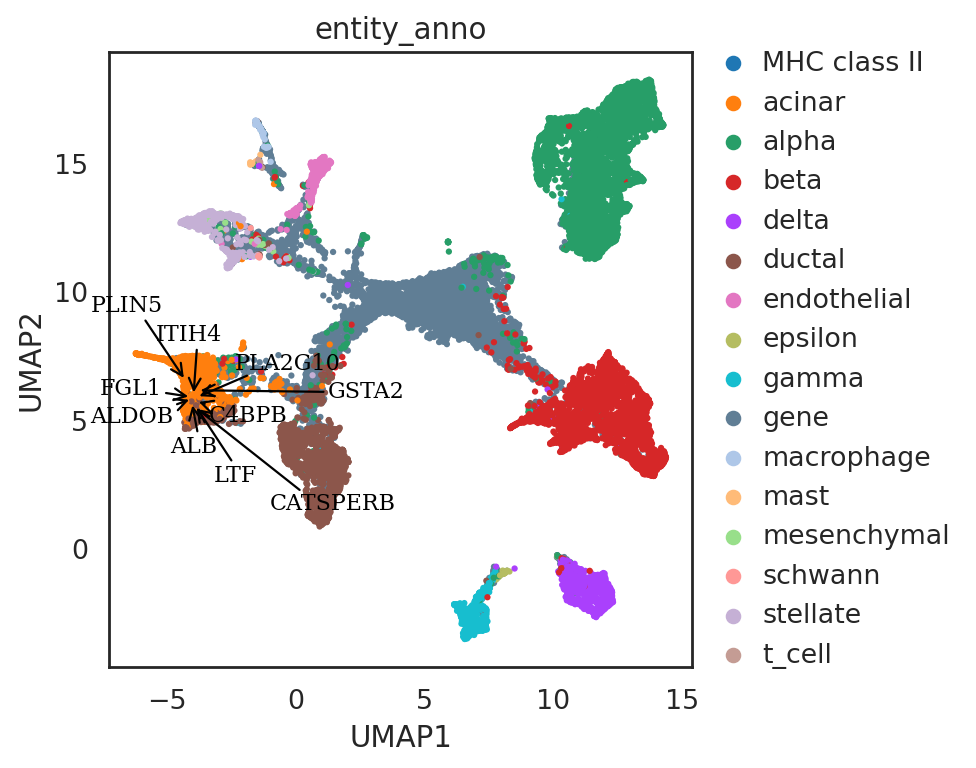
[ ]:
visualize these neighbor genes on UMAP of cells
[105]:
import numpy as np
adata_CG_concat = adata_CG_baron.concatenate([adata_CG_muraro,
adata_CG_segerstolpe,
adata_CG_wang,
adata_CG_xin],
join='outer',
fill_value=np.nan,
index_unique=None)
adata_CG_concat.obsm['X_umap'] = adata_all[adata_CG_concat.obs_names,].obsm['X_umap'].copy()
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/scipy/sparse/_index.py:125: SparseEfficiencyWarning: Changing the sparsity structure of a csc_matrix is expensive. lil_matrix is more efficient.
self._set_arrayXarray(i, j, x)
/data/pinello/SHARED_SOFTWARE/anaconda_latest/envs/hc_simba/lib/python3.7/site-packages/pandas/core/arrays/categorical.py:2487: FutureWarning: The `inplace` parameter in pandas.Categorical.remove_unused_categories is deprecated and will be removed in a future version.
res = method(*args, **kwargs)
[108]:
si.pl.umap(adata_CG_concat,
color=query_genes.index[:10],
drawing_order='sorted',
fig_ncol=5,
fig_size=(4,4))
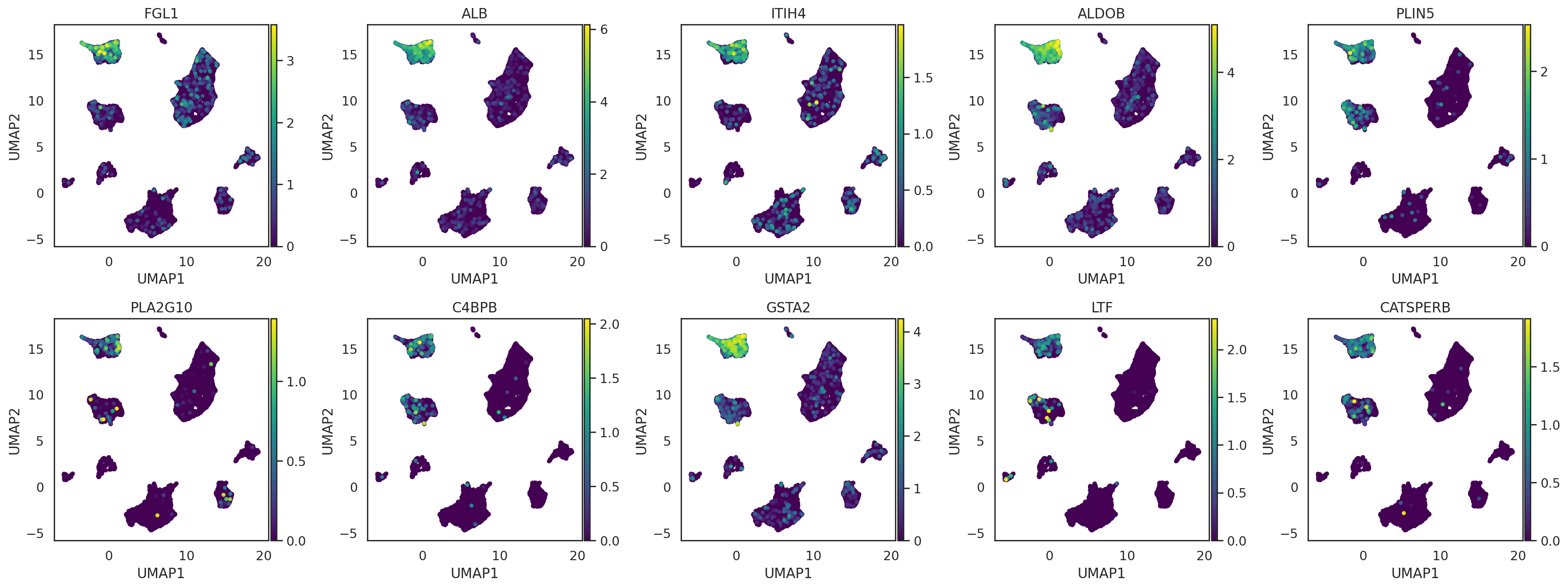
[ ]:
save results
[109]:
adata_CG_baron.write(os.path.join(workdir,'adata_CG_baron.h5ad'))
adata_CG_muraro.write(os.path.join(workdir,'adata_CG_muraro.h5ad'))
adata_CG_segerstolpe.write(os.path.join(workdir,'adata_CG_segerstolpe.h5ad'))
adata_CG_wang.write(os.path.join(workdir,'adata_CG_wang.h5ad'))
adata_CG_xin.write(os.path.join(workdir,'adata_CG_xin.h5ad'))
adata_C.write(os.path.join(workdir,'adata_C.h5ad'))
adata_C2.write(os.path.join(workdir,'adata_C2.h5ad'))
adata_C3.write(os.path.join(workdir,'adata_C3.h5ad'))
adata_C4.write(os.path.join(workdir,'adata_C4.h5ad'))
adata_C5.write(os.path.join(workdir,'adata_C5.h5ad'))
adata_G.write(os.path.join(workdir,'adata_G.h5ad'))
# adata_all.write(os.path.join(workdir,'adata_all.h5ad'))
# adata_all_CG.write(os.path.join(workdir,'adata_all_CG.h5ad'))
Read back anndata objects
adata_CG_baron = si.read_h5ad(os.path.join(workdir,'adata_CG_baron.h5ad'))
adata_CG_muraro = si.read_h5ad(os.path.join(workdir,'adata_CG_muraro.h5ad'))
adata_CG_segerstolpe = si.read_h5ad(os.path.join(workdir,'adata_CG_segerstolpe.h5ad'))
adata_CG_wang = si.read_h5ad(os.path.join(workdir,'adata_CG_wang.h5ad'))
adata_CG_xin = si.read_h5ad(os.path.join(workdir,'adata_CG_xin.h5ad'))
adata_C = si.read_h5ad(os.path.join(workdir,'adata_C.h5ad'))
adata_C2 = si.read_h5ad(os.path.join(workdir,'adata_C2.h5ad'))
adata_C3 = si.read_h5ad(os.path.join(workdir,'adata_C3.h5ad'))
adata_C4 = si.read_h5ad(os.path.join(workdir,'adata_C4.h5ad'))
adata_C5 = si.read_h5ad(os.path.join(workdir,'adata_C5.h5ad'))
adata_G = si.read_h5ad(os.path.join(workdir,'adata_G.h5ad'))
[ ]: