Generalized graph generation
In this tutorial, we will show how to use the generalized graph generation procedure by specifying the new parameter
list_adata in si.tl.gen_graph()-
si.tl.gen_graph() no longer requires explicit entity type specification. - Automatic matching of entities and relations between anndata objects. - Easy extension to incorporate new single-cell modalities.[1]:
import os
import simba as si
si.__version__
[1]:
'1.2'
[2]:
si.settings.set_figure_params(dpi=80,
style='white',
fig_size=[5,5],
rc={'image.cmap': 'viridis'})
[3]:
# make plots prettier
from matplotlib_inline.backend_inline import set_matplotlib_formats
set_matplotlib_formats('retina')
[ ]:
scRNA-seq analysis
[4]:
workdir = 'result_new_graph_generation/scRNAseq'
si.settings.set_workdir(workdir)
Saving results in: result_new_graph_generation/scRNAseq
[5]:
adata_CG = si.datasets.rna_10xpmbc3k()
si.pp.filter_genes(adata_CG,min_n_cells=3)
si.pp.cal_qc_rna(adata_CG)
si.pp.normalize(adata_CG,method='lib_size')
si.pp.log_transform(adata_CG)
si.pp.select_variable_genes(adata_CG, n_top_genes=2000)
Downloading data ...
rna_10xpmbc3k.h5ad: 21.5MB [00:03, 5.45MB/s]
Downloaded to result_new_graph_generation/scRNAseq/data.
Before filtering:
2700 cells, 32738 genes
Filter genes based on min_n_cells
After filtering out low-expressed genes:
2700 cells, 13714 genes
2000 variable genes are selected.
[6]:
# keep only variable genes
adata_CG_fltr = adata_CG[:, adata_CG.var['highly_variable']].copy()
si.tl.gen_graph(list_adata=[adata_CG_fltr],
copy=False,
add_edge_weights=True,
dirname='graph0')
`simba` does not exist in adata 0 in `list_adata`.`.X` is being used instead.
relation0: source: E0, destination: E1
#edges: 509430
Total number of edges: 509430
Writing graph file "pbg_graph.txt" to "result_new_graph_generation/scRNAseq/pbg/graph0" ...
Finished.
[7]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
dict_config['wd_interval'] = 10 # we usually set `wd_interval` to 10 for scRNA-seq datasets for a slower but finer training
dict_config['workers'] = 4 #The number of CPUs.
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model', use_edge_weights=True)
Auto-estimated weight decay is 0.069558
`.settings.pbg_params['wd']` has been updated to 0.069558
Converting input data ...
Edge weights are being used ...
[2022-10-10 15:29:26.273437] Using the 1 relation types given in the config
[2022-10-10 15:29:26.273982] Searching for the entities in the edge files...
[2022-10-10 15:29:27.014368] Entity type E0:
[2022-10-10 15:29:27.015074] - Found 2700 entities
[2022-10-10 15:29:27.015471] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 15:29:27.016342] - Left with 2700 entities
[2022-10-10 15:29:27.016984] - Shuffling them...
[2022-10-10 15:29:27.019567] Entity type E1:
[2022-10-10 15:29:27.020450] - Found 2000 entities
[2022-10-10 15:29:27.020810] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 15:29:27.021417] - Left with 2000 entities
[2022-10-10 15:29:27.021901] - Shuffling them...
[2022-10-10 15:29:27.023654] Preparing counts and dictionaries for entities and relation types:
[2022-10-10 15:29:27.024809] - Writing count of entity type E0 and partition 0
[2022-10-10 15:29:27.028415] - Writing count of entity type E1 and partition 0
[2022-10-10 15:29:27.031841] Preparing edge path result_new_graph_generation/scRNAseq/pbg/graph0/input/edge, out of the edges found in result_new_graph_generation/scRNAseq/pbg/graph0/pbg_graph.txt
using fast version
[2022-10-10 15:29:27.032810] Taking the fast train!
[2022-10-10 15:29:27.558752] - Processed 100000 edges so far...
[2022-10-10 15:29:28.043125] - Processed 200000 edges so far...
[2022-10-10 15:29:28.515778] - Processed 300000 edges so far...
[2022-10-10 15:29:29.080527] - Processed 400000 edges so far...
[2022-10-10 15:29:29.584989] - Processed 500000 edges so far...
[2022-10-10 15:29:30.366880] - Processed 509430 edges in total
Starting training ...
Finished
[8]:
si.pl.pbg_metrics(fig_ncol=1)
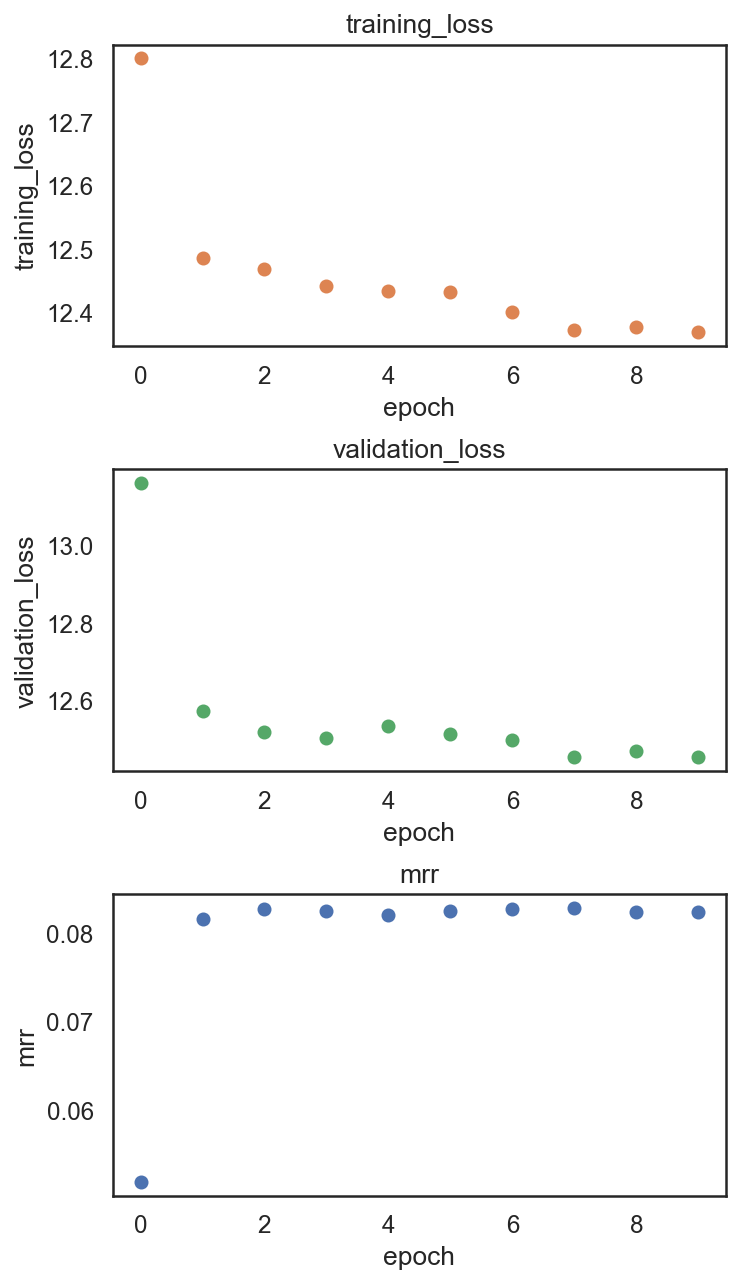
[9]:
palette_celltype={'B':'#1f77b4',
'CD4 T':'#ff7f0e',
'CD8 T':'#279e68',
'Dendritic':"#aa40fc",
'CD14 Monocytes':'#d62728',
'FCGR3A Monocytes':'#b5bd61',
'Megakaryocytes':'#e377c2',
'NK':'#8c564b'}
dict_adata = si.read_embedding()
adata_C = dict_adata['E0'] # embeddings for cells
adata_G = dict_adata['E1'] # embeddings for genes
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CG[adata_C.obs_names,:].obs['celltype'].copy()
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
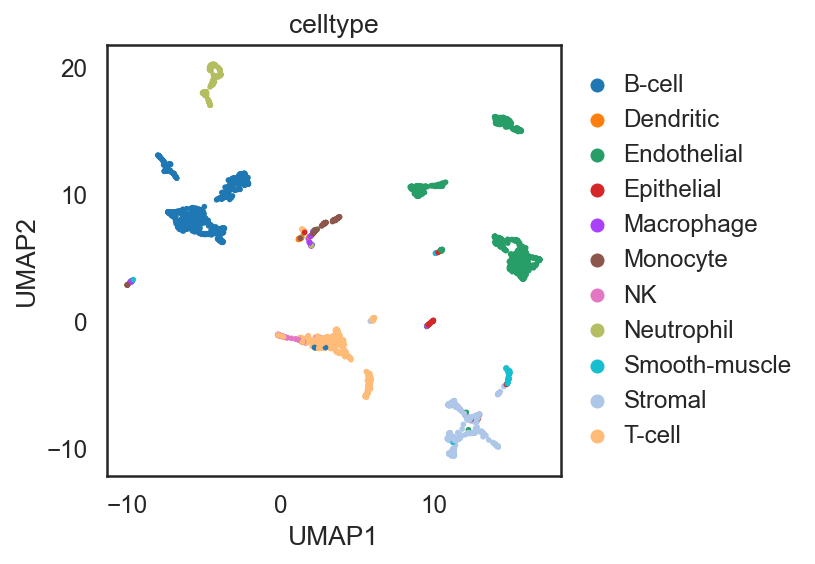
si.pl.umap(adata_C,color=['celltype'],
dict_palette={'celltype': palette_celltype},
fig_size=(6,4),
drawing_order='random')
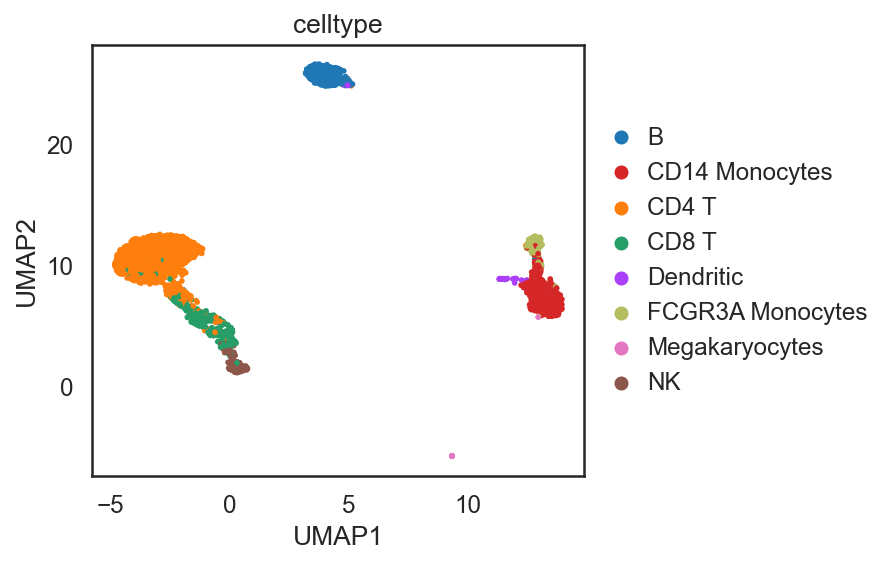
[ ]:
[ ]:
Batch correction (two batches)
[10]:
workdir = 'result_new_graph_generation/batch_correction_two'
si.settings.set_workdir(workdir)
Saving results in: result_new_graph_generation/batch_correction_two
[11]:
# mirowell-seq data
adata_CG_mi = si.datasets.rna_han2018()
# smart-seq data
adata_CG_sm = si.datasets.rna_tmc2018()
Downloading data ...
rna_han2018.h5ad: 24.6MB [00:04, 5.74MB/s]
Downloaded to result_new_graph_generation/batch_correction_two/data.
Downloading data ...
rna_tmc2018.h5ad: 50.9MB [00:07, 7.07MB/s]
Downloaded to result_new_graph_generation/batch_correction_two/data.
[12]:
adata_CG_mi
[12]:
AnnData object with n_obs × n_vars = 4239 × 15006
obs: 'nGene', 'nUMI', 'orig.ident', 'batch', 'ct', 'ct.orig', 'tissue', 'organ', 'percent.mito', 'batchlb', 'celltype'
[13]:
adata_CG_sm
[13]:
AnnData object with n_obs × n_vars = 2715 × 15006
obs: 'nGene', 'nUMI', 'orig.ident', 'batch', 'ct', 'ct.orig', 'tissue', 'organ', 'percent.mito', 'batchlb', 'celltype'
[14]:
# microwell-seq data
si.pp.filter_genes(adata_CG_mi,min_n_cells=3)
si.pp.cal_qc_rna(adata_CG_mi)
si.pp.normalize(adata_CG_mi,method='lib_size')
si.pp.log_transform(adata_CG_mi)
si.pp.select_variable_genes(adata_CG_mi, n_top_genes=3000)
Before filtering:
4239 cells, 15006 genes
Filter genes based on min_n_cells
After filtering out low-expressed genes:
4239 cells, 13662 genes
3000 variable genes are selected.
[15]:
# smart-seq2 data
si.pp.filter_genes(adata_CG_sm,min_n_cells=3)
si.pp.cal_qc_rna(adata_CG_sm)
si.pp.normalize(adata_CG_sm,method='lib_size')
si.pp.log_transform(adata_CG_sm)
si.pp.select_variable_genes(adata_CG_sm, n_top_genes=3000)
Before filtering:
2715 cells, 15006 genes
Filter genes based on min_n_cells
After filtering out low-expressed genes:
2715 cells, 14956 genes
3000 variable genes are selected.
[16]:
# infer edges between cells of different technologies
adata_CmiCsm = si.tl.infer_edges(adata_CG_mi, adata_CG_sm, n_components=20, k=20)
# edges can be futhere trimmed if needed. Here we keep all of them
si.tl.trim_edges(adata_CmiCsm, cutoff=0.5)
# The range of similary scores is 0-1. We multipled them by 10 to give higher weights to relation between cells
adata_CmiCsm.X = adata_CmiCsm.X*10
#shared features: 2969
Performing randomized SVD ...
Searching for mutual nearest neighbors ...
27952 edges are selected
27952 edges are selected
/Users/huidong/anaconda3/envs/env_simba_dev/lib/python3.10/site-packages/simba/tools/_integration.py:142: FutureWarning: X.dtype being converted to np.float32 from float64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
adata_ref_query = ad.AnnData(X=sim_ref_query,
[17]:
adata_CmiCsm
[17]:
AnnData object with n_obs × n_vars = 4239 × 2715
obs: 'nGene', 'nUMI', 'orig.ident', 'batch', 'ct', 'ct.orig', 'tissue', 'organ', 'percent.mito', 'batchlb', 'celltype', 'n_counts', 'n_genes', 'pct_genes', 'pct_mt'
var: 'nGene', 'nUMI', 'orig.ident', 'batch', 'ct', 'ct.orig', 'tissue', 'organ', 'percent.mito', 'batchlb', 'celltype', 'n_counts', 'n_genes', 'pct_genes', 'pct_mt'
obsm: 'svd'
varm: 'svd'
layers: 'simba'
[18]:
# keep only variable genes
adata_CG_mi_fltr = adata_CG_mi[:, adata_CG_mi.var['highly_variable']].copy()
adata_CG_sm_fltr = adata_CG_sm[:, adata_CG_sm.var['highly_variable']].copy()
si.tl.gen_graph(list_adata=[adata_CG_mi_fltr, adata_CG_sm_fltr, adata_CmiCsm],
copy=False,
layer=None,
dirname='graph2')
relation0: source: E0, destination: E1
#edges: 550497
relation1: source: E2, destination: E1
#edges: 773587
relation2: source: E0, destination: E2
#edges: 27952
Total number of edges: 1352036
Writing graph file "pbg_graph.txt" to "result_new_graph_generation/batch_correction_two/pbg/graph2" ...
Finished.
[19]:
si.settings.pbg_params
[19]:
{'entity_path': 'result_new_graph_generation/batch_correction_two/pbg/graph2/input/entity',
'edge_paths': ['result_new_graph_generation/batch_correction_two/pbg/graph2/input/edge'],
'checkpoint_path': '',
'entities': {'E0': {'num_partitions': 1},
'E1': {'num_partitions': 1},
'E2': {'num_partitions': 1}},
'relations': [{'name': 'r0',
'lhs': 'E0',
'rhs': 'E1',
'operator': 'none',
'weight': 1.0},
{'name': 'r1', 'lhs': 'E2', 'rhs': 'E1', 'operator': 'none', 'weight': 1.0},
{'name': 'r2', 'lhs': 'E0', 'rhs': 'E2', 'operator': 'none', 'weight': 1.0}],
'dynamic_relations': False,
'dimension': 50,
'global_emb': False,
'comparator': 'dot',
'num_epochs': 10,
'workers': 4,
'num_batch_negs': 50,
'num_uniform_negs': 50,
'loss_fn': 'softmax',
'lr': 0.1,
'early_stopping': False,
'regularization_coef': 0.0,
'wd': 0.0,
'wd_interval': 50,
'eval_fraction': 0.05,
'eval_num_batch_negs': 50,
'eval_num_uniform_negs': 50,
'checkpoint_preservation_interval': None}
[20]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
# dict_config['wd'] = 0.026209
dict_config['workers'] = 4
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model', use_edge_weights=True)
Auto-estimated weight decay is 0.026209
`.settings.pbg_params['wd']` has been updated to 0.026209
Converting input data ...
Edge weights are being used ...
[2022-10-10 15:30:17.233560] Using the 3 relation types given in the config
[2022-10-10 15:30:17.234069] Searching for the entities in the edge files...
[2022-10-10 15:30:19.057184] Entity type E0:
[2022-10-10 15:30:19.057731] - Found 4239 entities
[2022-10-10 15:30:19.058159] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 15:30:19.059129] - Left with 4239 entities
[2022-10-10 15:30:19.059906] - Shuffling them...
[2022-10-10 15:30:19.062890] Entity type E1:
[2022-10-10 15:30:19.063783] - Found 4620 entities
[2022-10-10 15:30:19.064333] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 15:30:19.065608] - Left with 4620 entities
[2022-10-10 15:30:19.066450] - Shuffling them...
[2022-10-10 15:30:19.069455] Entity type E2:
[2022-10-10 15:30:19.070294] - Found 2715 entities
[2022-10-10 15:30:19.070697] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 15:30:19.071529] - Left with 2715 entities
[2022-10-10 15:30:19.072205] - Shuffling them...
[2022-10-10 15:30:19.074476] Preparing counts and dictionaries for entities and relation types:
[2022-10-10 15:30:19.075873] - Writing count of entity type E0 and partition 0
[2022-10-10 15:30:19.081341] - Writing count of entity type E1 and partition 0
[2022-10-10 15:30:19.085846] - Writing count of entity type E2 and partition 0
[2022-10-10 15:30:19.090744] Preparing edge path result_new_graph_generation/batch_correction_two/pbg/graph2/input/edge, out of the edges found in result_new_graph_generation/batch_correction_two/pbg/graph2/pbg_graph.txt
using fast version
[2022-10-10 15:30:19.091617] Taking the fast train!
[2022-10-10 15:30:19.573987] - Processed 100000 edges so far...
[2022-10-10 15:30:20.067008] - Processed 200000 edges so far...
[2022-10-10 15:30:20.533667] - Processed 300000 edges so far...
[2022-10-10 15:30:21.004497] - Processed 400000 edges so far...
[2022-10-10 15:30:21.464137] - Processed 500000 edges so far...
[2022-10-10 15:30:21.935342] - Processed 600000 edges so far...
[2022-10-10 15:30:22.390411] - Processed 700000 edges so far...
[2022-10-10 15:30:22.867359] - Processed 800000 edges so far...
[2022-10-10 15:30:23.333513] - Processed 900000 edges so far...
[2022-10-10 15:30:23.812747] - Processed 1000000 edges so far...
[2022-10-10 15:30:24.270701] - Processed 1100000 edges so far...
[2022-10-10 15:30:24.735069] - Processed 1200000 edges so far...
[2022-10-10 15:30:25.206952] - Processed 1300000 edges so far...
[2022-10-10 15:30:27.197654] - Processed 1352036 edges in total
Starting training ...
Finished
[21]:
si.pl.pbg_metrics(fig_ncol=1)
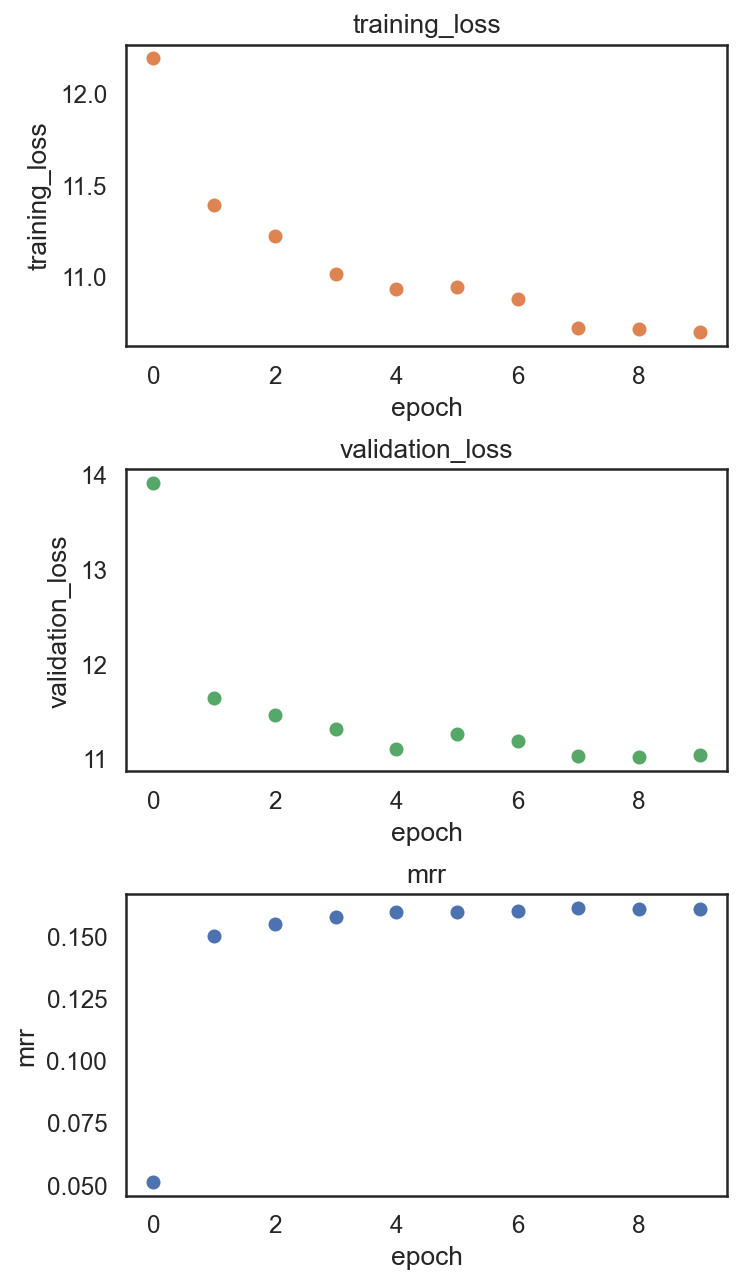
[22]:
dict_adata = si.read_embedding()
adata_C = dict_adata['E0'] # embeddings of cells from mirowell-seq data
adata_C2 = dict_adata['E2'] # embeddings for cells from smart-seq data
adata_G = dict_adata['E1'] # embeddings for genes
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CG_mi[adata_C.obs_names,:].obs['celltype'].copy()
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
## Add annotation of celltypes (optional)
adata_C2.obs['celltype'] = adata_CG_sm[adata_C2.obs_names,:].obs['celltype'].copy()
si.tl.umap(adata_C2,n_neighbors=15,n_components=2)
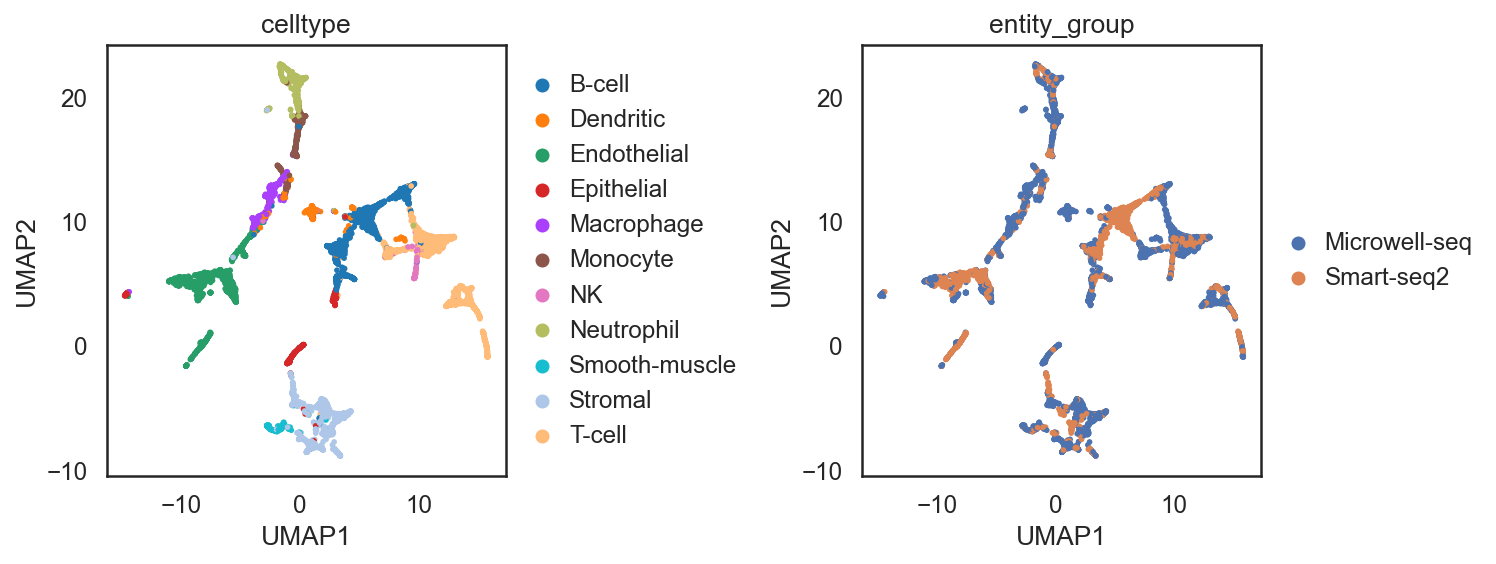
adata_all = si.tl.embed(adata_ref=adata_C,list_adata_query=[adata_C2])
## add annotations of two batches
adata_all.obs['entity_group'] = ""
adata_all.obs.loc[adata_C.obs_names, 'entity_group'] = "Microwell-seq"
adata_all.obs.loc[adata_C2.obs_names, 'entity_group'] = "Smart-seq2"
adata_all.obs.head()
si.tl.umap(adata_all,n_neighbors=15,n_components=2)
si.pl.umap(adata_C,
color=['celltype'],
fig_size=(5.5, 4),
drawing_order='random')
si.pl.umap(adata_C2,
color=['celltype'],
fig_size=(5.5, 4),
drawing_order='random')
si.pl.umap(adata_all,color=['celltype','entity_group'],
drawing_order='random',
fig_size=(5,4))
Performing softmax transformation for query data 0;
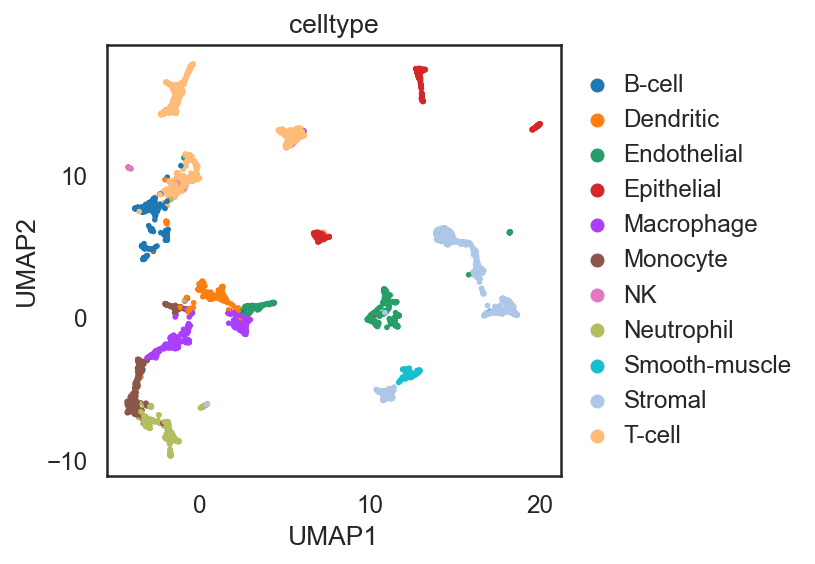
[ ]: