Use edge weigthts
In this tutorial, we will show how to incorporate weights into the graph (add_edge_weights=True) and enable these weights for training (use_edge_weights=True)
[1]:
import os
import simba as si
si.__version__
[1]:
'1.2'
[2]:
workdir = 'result_simba_edge_weights'
si.settings.set_workdir(workdir)
Saving results in: result_simba_edge_weights
[3]:
si.settings.set_figure_params(dpi=80,
style='white',
fig_size=[5,5],
rc={'image.cmap': 'viridis'})
[4]:
# make plots prettier
from matplotlib_inline.backend_inline import set_matplotlib_formats
set_matplotlib_formats('retina')
[5]:
adata_CG = si.datasets.rna_10xpmbc3k()
Downloading data ...
rna_10xpmbc3k.h5ad: 21.5MB [00:03, 5.60MB/s]
Downloaded to result_simba_edge_weights/data.
[6]:
adata_CG
[6]:
AnnData object with n_obs × n_vars = 2700 × 32738
obs: 'celltype'
var: 'gene_ids'
[7]:
si.pp.filter_genes(adata_CG,min_n_cells=3)
Before filtering:
2700 cells, 32738 genes
Filter genes based on min_n_cells
After filtering out low-expressed genes:
2700 cells, 13714 genes
[8]:
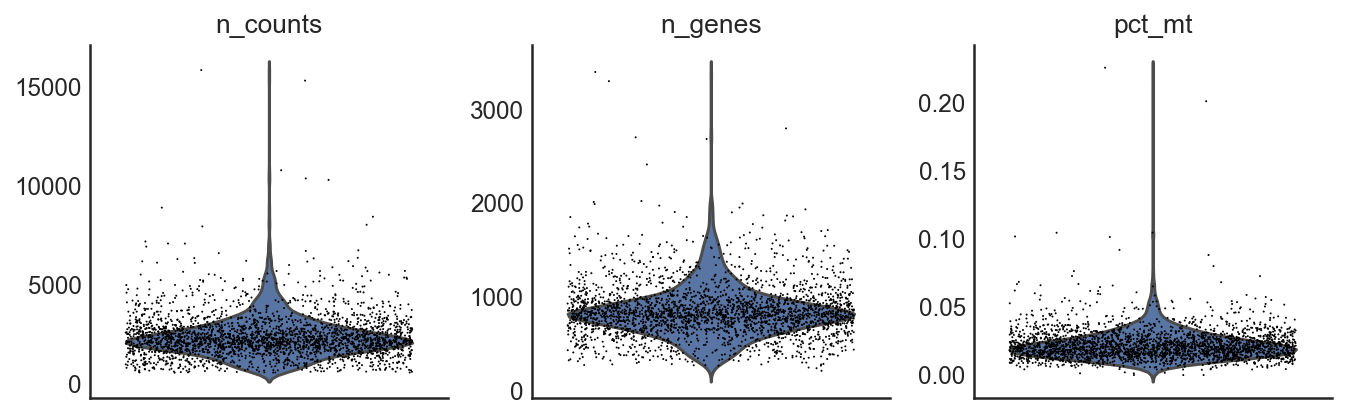
si.pp.cal_qc_rna(adata_CG)
si.pl.violin(adata_CG,list_obs=['n_counts','n_genes','pct_mt'])
[9]:
si.pp.normalize(adata_CG,method='lib_size')
si.pp.log_transform(adata_CG)
[10]:
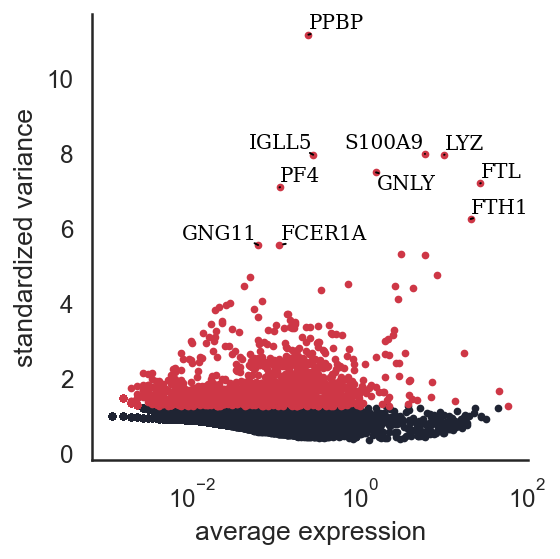
si.pp.select_variable_genes(adata_CG, n_top_genes=2000)
si.pl.variable_genes(adata_CG,show_texts=True)
2000 variable genes are selected.
[11]:
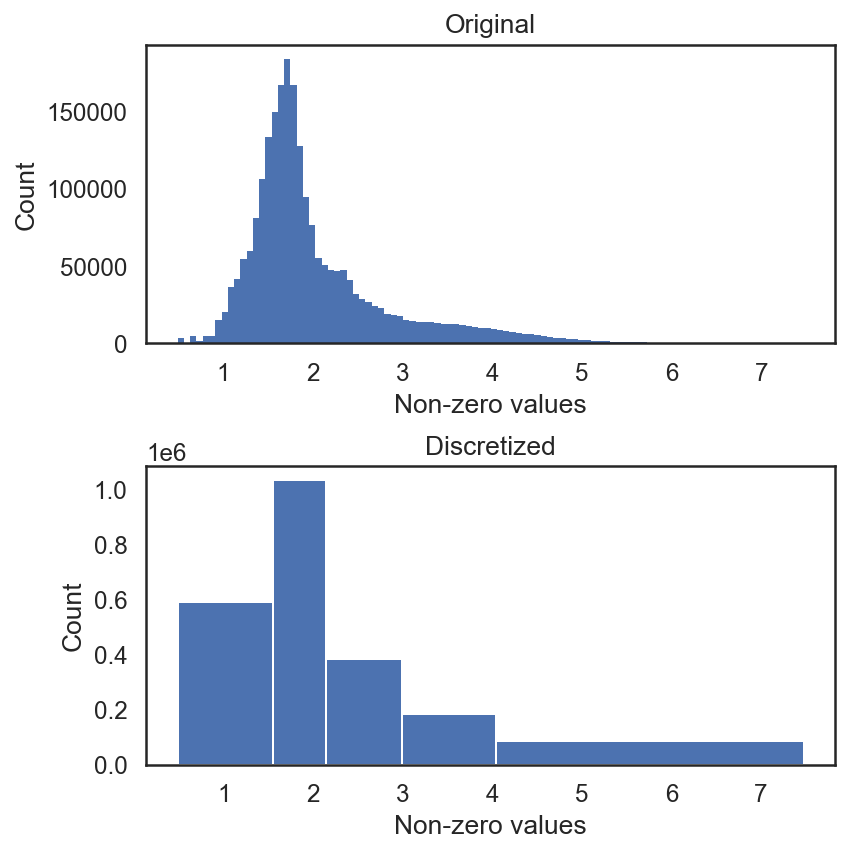
si.tl.discretize(adata_CG,n_bins=5)
si.pl.discretize(adata_CG,kde=False)
[ ]:
Using discretized gene expression - all genes
[12]:
si.tl.gen_graph(list_CG=[adata_CG],
copy=False,
use_highly_variable=False,
layer='simba',
add_edge_weights=False,
dirname='graph0')
relation0: source: C, destination: G
#edges: 590134
relation1: source: C, destination: G
#edges: 1034817
relation2: source: C, destination: G
#edges: 384939
relation3: source: C, destination: G
#edges: 185485
relation4: source: C, destination: G
#edges: 87601
Total number of edges: 2282976
Writing graph file "pbg_graph.txt" to "result_simba_edge_weights/pbg/graph0" ...
Finished.
[13]:
si.settings.pbg_params
[13]:
{'entity_path': 'result_simba_edge_weights/pbg/graph0/input/entity',
'edge_paths': ['result_simba_edge_weights/pbg/graph0/input/edge'],
'checkpoint_path': '',
'entities': {'C': {'num_partitions': 1}, 'G': {'num_partitions': 1}},
'relations': [{'name': 'r0',
'lhs': 'C',
'rhs': 'G',
'operator': 'none',
'weight': 1.0},
{'name': 'r1', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 2.0},
{'name': 'r2', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 3.0},
{'name': 'r3', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 4.0},
{'name': 'r4', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 5.0}],
'dynamic_relations': False,
'dimension': 50,
'global_emb': False,
'comparator': 'dot',
'num_epochs': 10,
'workers': 4,
'num_batch_negs': 50,
'num_uniform_negs': 50,
'loss_fn': 'softmax',
'lr': 0.1,
'early_stopping': False,
'regularization_coef': 0.0,
'wd': 0.0,
'wd_interval': 50,
'eval_fraction': 0.05,
'eval_num_batch_negs': 50,
'eval_num_uniform_negs': 50,
'checkpoint_preservation_interval': None}
[14]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
# dict_config['wd'] = 0.015521
dict_config['wd_interval'] = 10 # we usually set `wd_interval` to 10 for scRNA-seq datasets for a slower but finer training
dict_config['workers'] = 4 #The number of CPUs.
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model')
Auto-estimated weight decay is 0.015521
`.settings.pbg_params['wd']` has been updated to 0.015521
Converting input data ...
[2022-10-10 13:43:03.453400] Using the 5 relation types given in the config
[2022-10-10 13:43:03.453905] Searching for the entities in the edge files...
[2022-10-10 13:43:06.243132] Entity type C:
[2022-10-10 13:43:06.243681] - Found 2700 entities
[2022-10-10 13:43:06.244079] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:43:06.244885] - Left with 2700 entities
[2022-10-10 13:43:06.245435] - Shuffling them...
[2022-10-10 13:43:06.247549] Entity type G:
[2022-10-10 13:43:06.248129] - Found 13714 entities
[2022-10-10 13:43:06.248461] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:43:06.250226] - Left with 13714 entities
[2022-10-10 13:43:06.250876] - Shuffling them...
[2022-10-10 13:43:06.259329] Preparing counts and dictionaries for entities and relation types:
[2022-10-10 13:43:06.260700] - Writing count of entity type C and partition 0
[2022-10-10 13:43:06.264633] - Writing count of entity type G and partition 0
[2022-10-10 13:43:06.272220] Preparing edge path result_simba_edge_weights/pbg/graph0/input/edge, out of the edges found in result_simba_edge_weights/pbg/graph0/pbg_graph.txt
using fast version
[2022-10-10 13:43:06.272903] Taking the fast train!
[2022-10-10 13:43:06.731291] - Processed 100000 edges so far...
[2022-10-10 13:43:07.196747] - Processed 200000 edges so far...
[2022-10-10 13:43:07.699447] - Processed 300000 edges so far...
[2022-10-10 13:43:08.199365] - Processed 400000 edges so far...
[2022-10-10 13:43:08.673068] - Processed 500000 edges so far...
[2022-10-10 13:43:09.150632] - Processed 600000 edges so far...
[2022-10-10 13:43:09.614157] - Processed 700000 edges so far...
[2022-10-10 13:43:10.094486] - Processed 800000 edges so far...
[2022-10-10 13:43:10.555303] - Processed 900000 edges so far...
[2022-10-10 13:43:11.048349] - Processed 1000000 edges so far...
[2022-10-10 13:43:11.544103] - Processed 1100000 edges so far...
[2022-10-10 13:43:12.053794] - Processed 1200000 edges so far...
[2022-10-10 13:43:12.541551] - Processed 1300000 edges so far...
[2022-10-10 13:43:13.064220] - Processed 1400000 edges so far...
[2022-10-10 13:43:13.527216] - Processed 1500000 edges so far...
[2022-10-10 13:43:13.981077] - Processed 1600000 edges so far...
[2022-10-10 13:43:14.459946] - Processed 1700000 edges so far...
[2022-10-10 13:43:14.970111] - Processed 1800000 edges so far...
[2022-10-10 13:43:15.444465] - Processed 1900000 edges so far...
[2022-10-10 13:43:15.988276] - Processed 2000000 edges so far...
[2022-10-10 13:43:16.434461] - Processed 2100000 edges so far...
[2022-10-10 13:43:16.889269] - Processed 2200000 edges so far...
[2022-10-10 13:43:19.983264] - Processed 2282976 edges in total
Starting training ...
Finished
[15]:
si.pl.pbg_metrics(fig_ncol=1)
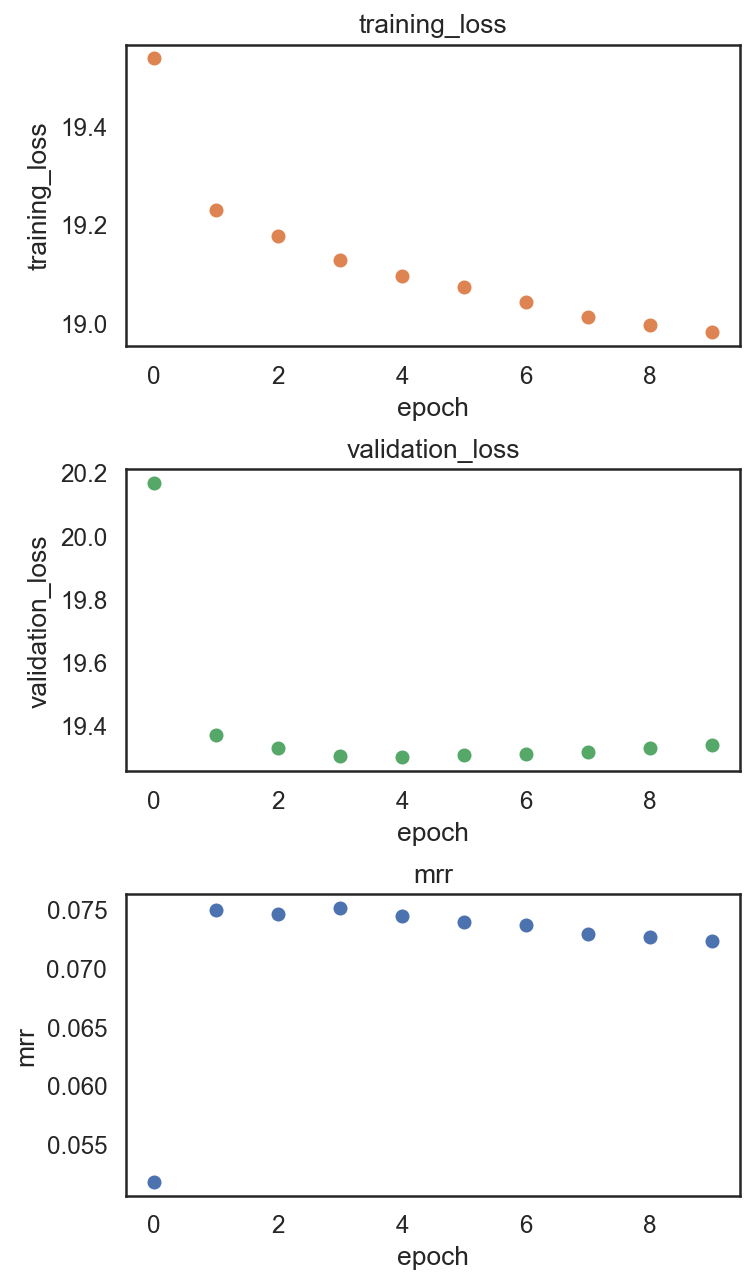
[16]:
palette_celltype={'B':'#1f77b4',
'CD4 T':'#ff7f0e',
'CD8 T':'#279e68',
'Dendritic':"#aa40fc",
'CD14 Monocytes':'#d62728',
'FCGR3A Monocytes':'#b5bd61',
'Megakaryocytes':'#e377c2',
'NK':'#8c564b'}
dict_adata = si.read_embedding()
adata_C = dict_adata['C'] # embeddings for cells
adata_G = dict_adata['G'] # embeddings for genes
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CG[adata_C.obs_names,:].obs['celltype'].copy()
adata_C
[16]:
AnnData object with n_obs × n_vars = 2700 × 50
obs: 'celltype'
[17]:
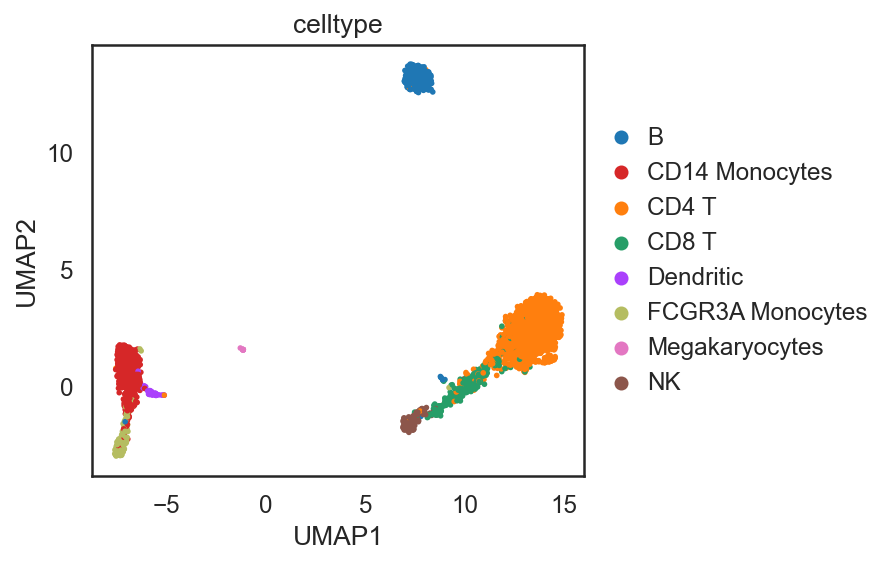
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
si.pl.umap(adata_C,color=['celltype'],
dict_palette={'celltype': palette_celltype},
fig_size=(6,4),
drawing_order='random')
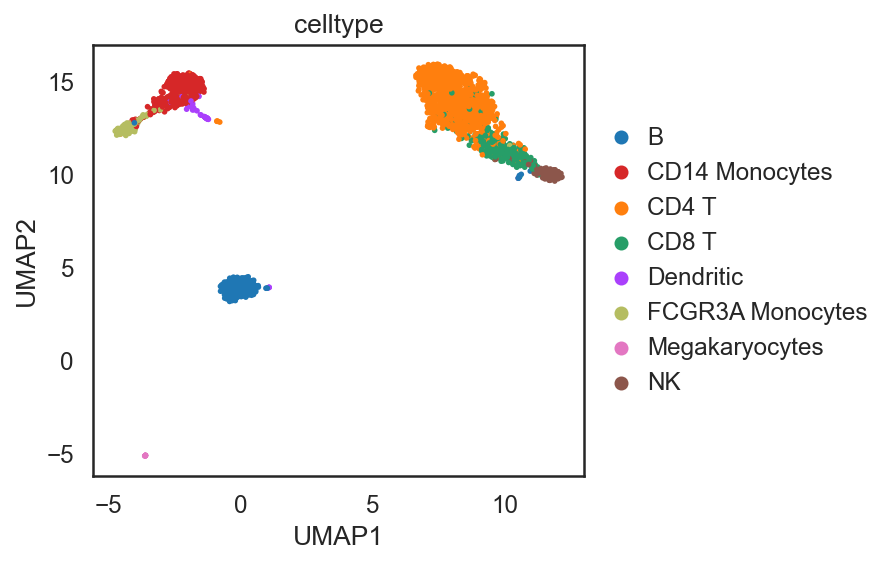
[ ]:
Using discretized gene expression - only variable genes
[18]:
si.tl.gen_graph(list_CG=[adata_CG],
copy=False,
use_highly_variable=True,
layer='simba',
add_edge_weights=False,
dirname='graph1')
relation0: source: C, destination: G
#edges: 138727
relation1: source: C, destination: G
#edges: 243405
relation2: source: C, destination: G
#edges: 82172
relation3: source: C, destination: G
#edges: 27876
relation4: source: C, destination: G
#edges: 17250
Total number of edges: 509430
Writing graph file "pbg_graph.txt" to "result_simba_edge_weights/pbg/graph1" ...
Finished.
[19]:
si.settings.pbg_params
[19]:
{'entity_path': 'result_simba_edge_weights/pbg/graph1/input/entity',
'edge_paths': ['result_simba_edge_weights/pbg/graph1/input/edge'],
'checkpoint_path': 'result_simba_edge_weights/pbg/graph0/model',
'entities': {'C': {'num_partitions': 1}, 'G': {'num_partitions': 1}},
'relations': [{'name': 'r0',
'lhs': 'C',
'rhs': 'G',
'operator': 'none',
'weight': 1.0},
{'name': 'r1', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 2.0},
{'name': 'r2', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 3.0},
{'name': 'r3', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 4.0},
{'name': 'r4', 'lhs': 'C', 'rhs': 'G', 'operator': 'none', 'weight': 5.0}],
'dynamic_relations': False,
'dimension': 50,
'global_emb': False,
'comparator': 'dot',
'num_epochs': 10,
'workers': 4,
'num_batch_negs': 50,
'num_uniform_negs': 50,
'loss_fn': 'softmax',
'lr': 0.1,
'early_stopping': False,
'regularization_coef': 0.0,
'wd': 0.015521,
'wd_interval': 50,
'eval_fraction': 0.05,
'eval_num_batch_negs': 50,
'eval_num_uniform_negs': 50,
'checkpoint_preservation_interval': None}
[20]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
dict_config['wd_interval'] = 10 # we usually set `wd_interval` to 10 for scRNA-seq datasets for a slower but finer training
dict_config['workers'] = 4 #The number of CPUs.
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model')
Auto-estimated weight decay is 0.069558
`.settings.pbg_params['wd']` has been updated to 0.069558
Converting input data ...
[2022-10-10 13:44:46.615190] Using the 5 relation types given in the config
[2022-10-10 13:44:46.615748] Searching for the entities in the edge files...
[2022-10-10 13:44:47.245409] Entity type C:
[2022-10-10 13:44:47.246209] - Found 2700 entities
[2022-10-10 13:44:47.246714] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:44:47.247570] - Left with 2700 entities
[2022-10-10 13:44:47.247964] - Shuffling them...
[2022-10-10 13:44:47.249921] Entity type G:
[2022-10-10 13:44:47.250608] - Found 2000 entities
[2022-10-10 13:44:47.250971] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:44:47.251637] - Left with 2000 entities
[2022-10-10 13:44:47.252242] - Shuffling them...
[2022-10-10 13:44:47.253708] Preparing counts and dictionaries for entities and relation types:
[2022-10-10 13:44:47.254991] - Writing count of entity type C and partition 0
[2022-10-10 13:44:47.258398] - Writing count of entity type G and partition 0
[2022-10-10 13:44:47.260535] Preparing edge path result_simba_edge_weights/pbg/graph1/input/edge, out of the edges found in result_simba_edge_weights/pbg/graph1/pbg_graph.txt
using fast version
[2022-10-10 13:44:47.261206] Taking the fast train!
[2022-10-10 13:44:47.724024] - Processed 100000 edges so far...
[2022-10-10 13:44:48.175073] - Processed 200000 edges so far...
[2022-10-10 13:44:48.618815] - Processed 300000 edges so far...
[2022-10-10 13:44:49.075198] - Processed 400000 edges so far...
[2022-10-10 13:44:49.545616] - Processed 500000 edges so far...
[2022-10-10 13:44:50.091471] - Processed 509430 edges in total
Starting training ...
Finished
[21]:
si.pl.pbg_metrics(fig_ncol=1)
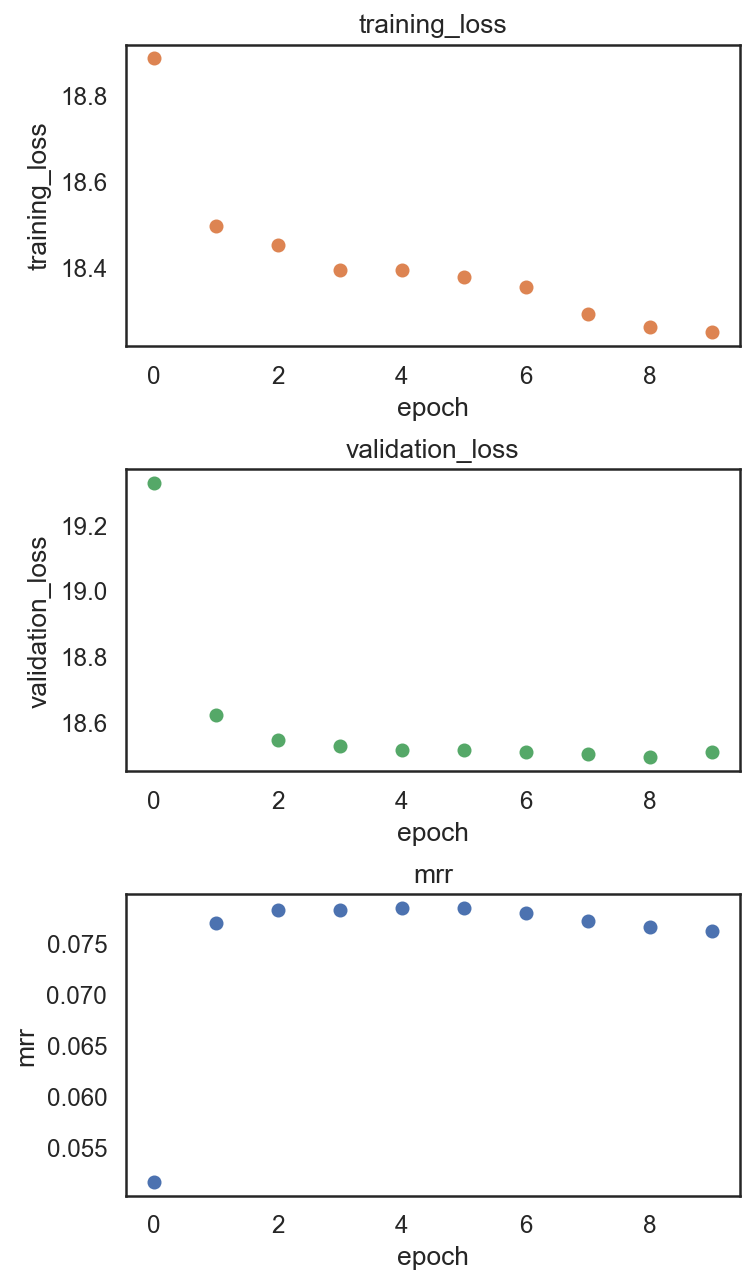
[22]:
palette_celltype={'B':'#1f77b4',
'CD4 T':'#ff7f0e',
'CD8 T':'#279e68',
'Dendritic':"#aa40fc",
'CD14 Monocytes':'#d62728',
'FCGR3A Monocytes':'#b5bd61',
'Megakaryocytes':'#e377c2',
'NK':'#8c564b'}
dict_adata = si.read_embedding()
adata_C = dict_adata['C'] # embeddings for cells
adata_G = dict_adata['G'] # embeddings for genes
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CG[adata_C.obs_names,:].obs['celltype'].copy()
adata_C
[22]:
AnnData object with n_obs × n_vars = 2700 × 50
obs: 'celltype'
[23]:
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
si.pl.umap(adata_C,color=['celltype'],
dict_palette={'celltype': palette_celltype},
fig_size=(6,4),
drawing_order='random')
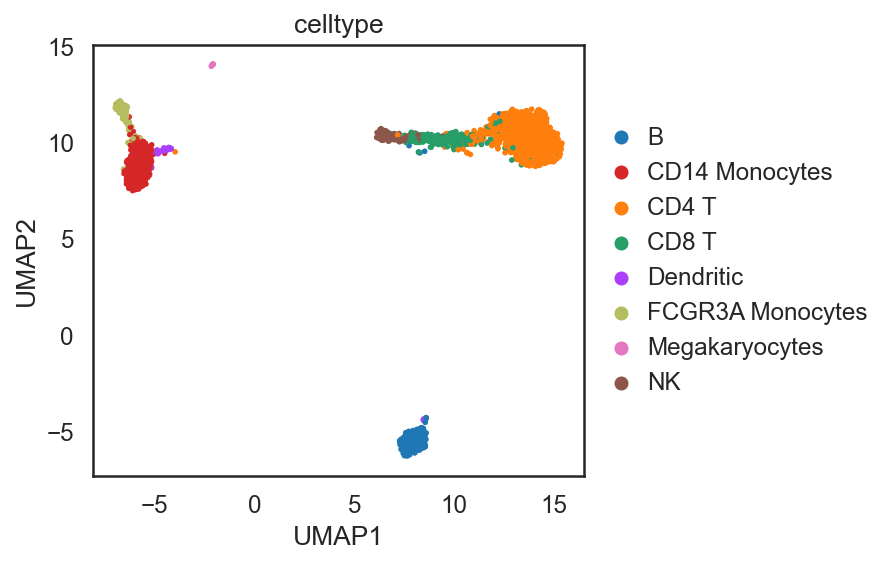
[ ]:
Using edge weights (raw gene expression) - all genes
[24]:
si.tl.gen_graph(list_CG=[adata_CG],
copy=False,
use_highly_variable=False,
layer=None,
add_edge_weights=True,
dirname='graph2')
relation0: source: C, destination: G
#edges: 2282976
Total number of edges: 2282976
Writing graph file "pbg_graph.txt" to "result_simba_edge_weights/pbg/graph2" ...
Finished.
[25]:
si.settings.pbg_params
[25]:
{'entity_path': 'result_simba_edge_weights/pbg/graph2/input/entity',
'edge_paths': ['result_simba_edge_weights/pbg/graph2/input/edge'],
'checkpoint_path': 'result_simba_edge_weights/pbg/graph1/model',
'entities': {'C': {'num_partitions': 1}, 'G': {'num_partitions': 1}},
'relations': [{'name': 'r0',
'lhs': 'C',
'rhs': 'G',
'operator': 'none',
'weight': 1.0}],
'dynamic_relations': False,
'dimension': 50,
'global_emb': False,
'comparator': 'dot',
'num_epochs': 10,
'workers': 4,
'num_batch_negs': 50,
'num_uniform_negs': 50,
'loss_fn': 'softmax',
'lr': 0.1,
'early_stopping': False,
'regularization_coef': 0.0,
'wd': 0.069558,
'wd_interval': 50,
'eval_fraction': 0.05,
'eval_num_batch_negs': 50,
'eval_num_uniform_negs': 50,
'checkpoint_preservation_interval': None}
[26]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
dict_config['wd_interval'] = 10 # we usually set `wd_interval` to 10 for scRNA-seq datasets for a slower but finer training
dict_config['workers'] = 4 #The number of CPUs.
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model', use_edge_weights=True)
Auto-estimated weight decay is 0.015521
`.settings.pbg_params['wd']` has been updated to 0.015521
Converting input data ...
Edge weights are being used ...
[2022-10-10 13:47:20.130429] Using the 1 relation types given in the config
[2022-10-10 13:47:20.131066] Searching for the entities in the edge files...
[2022-10-10 13:47:23.633055] Entity type C:
[2022-10-10 13:47:23.633609] - Found 2700 entities
[2022-10-10 13:47:23.633978] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:47:23.634907] - Left with 2700 entities
[2022-10-10 13:47:23.635662] - Shuffling them...
[2022-10-10 13:47:23.637645] Entity type G:
[2022-10-10 13:47:23.638327] - Found 13714 entities
[2022-10-10 13:47:23.638688] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:47:23.641269] - Left with 13714 entities
[2022-10-10 13:47:23.641881] - Shuffling them...
[2022-10-10 13:47:23.654717] Preparing counts and dictionaries for entities and relation types:
[2022-10-10 13:47:23.656340] - Writing count of entity type C and partition 0
[2022-10-10 13:47:23.659567] - Writing count of entity type G and partition 0
[2022-10-10 13:47:23.672482] Preparing edge path result_simba_edge_weights/pbg/graph2/input/edge, out of the edges found in result_simba_edge_weights/pbg/graph2/pbg_graph.txt
using fast version
[2022-10-10 13:47:23.673757] Taking the fast train!
[2022-10-10 13:47:24.203203] - Processed 100000 edges so far...
[2022-10-10 13:47:24.687041] - Processed 200000 edges so far...
[2022-10-10 13:47:25.188701] - Processed 300000 edges so far...
[2022-10-10 13:47:25.682914] - Processed 400000 edges so far...
[2022-10-10 13:47:26.187692] - Processed 500000 edges so far...
[2022-10-10 13:47:26.690817] - Processed 600000 edges so far...
[2022-10-10 13:47:27.297040] - Processed 700000 edges so far...
[2022-10-10 13:47:27.926251] - Processed 800000 edges so far...
[2022-10-10 13:47:28.443981] - Processed 900000 edges so far...
[2022-10-10 13:47:28.980157] - Processed 1000000 edges so far...
[2022-10-10 13:47:29.501237] - Processed 1100000 edges so far...
[2022-10-10 13:47:30.028912] - Processed 1200000 edges so far...
[2022-10-10 13:47:30.522380] - Processed 1300000 edges so far...
[2022-10-10 13:47:31.095499] - Processed 1400000 edges so far...
[2022-10-10 13:47:31.593399] - Processed 1500000 edges so far...
[2022-10-10 13:47:32.110170] - Processed 1600000 edges so far...
[2022-10-10 13:47:32.622942] - Processed 1700000 edges so far...
[2022-10-10 13:47:33.144220] - Processed 1800000 edges so far...
[2022-10-10 13:47:33.635787] - Processed 1900000 edges so far...
[2022-10-10 13:47:34.151326] - Processed 2000000 edges so far...
[2022-10-10 13:47:34.648824] - Processed 2100000 edges so far...
[2022-10-10 13:47:35.203821] - Processed 2200000 edges so far...
[2022-10-10 13:47:38.466443] - Processed 2282976 edges in total
Starting training ...
Finished
[27]:
si.pl.pbg_metrics(fig_ncol=1)
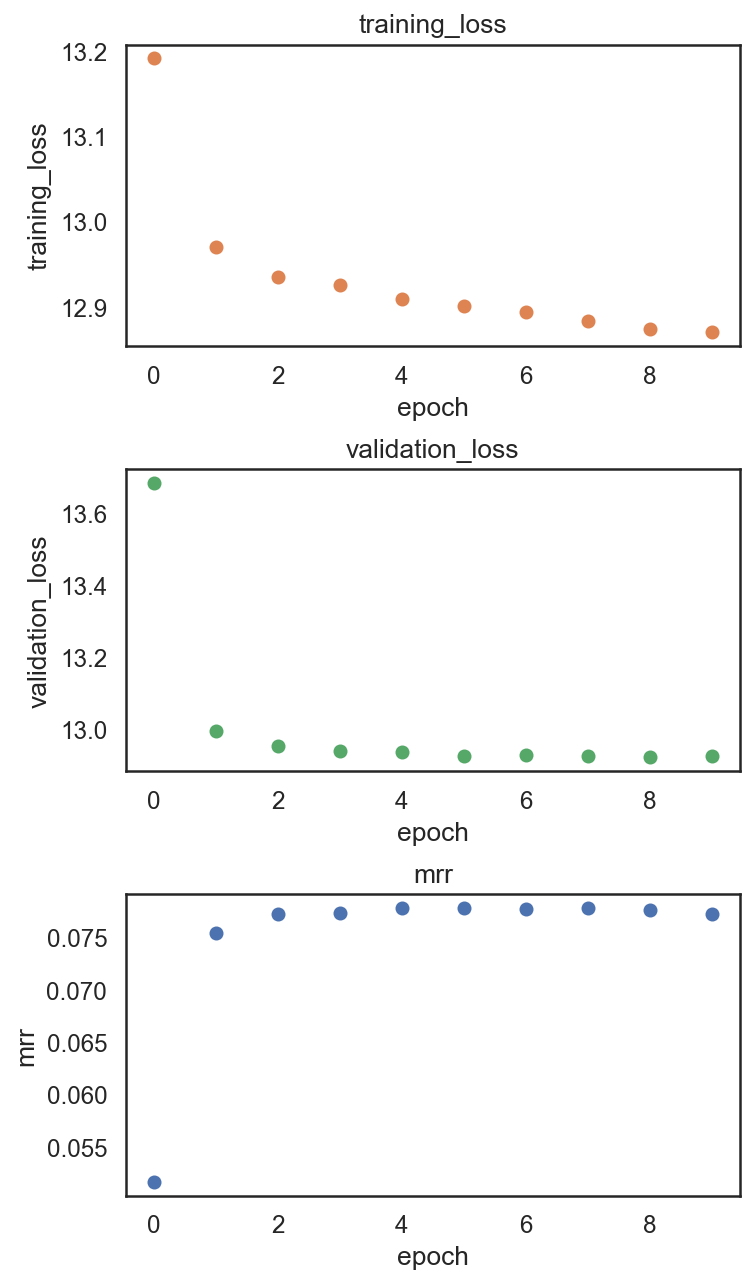
[28]:
palette_celltype={'B':'#1f77b4',
'CD4 T':'#ff7f0e',
'CD8 T':'#279e68',
'Dendritic':"#aa40fc",
'CD14 Monocytes':'#d62728',
'FCGR3A Monocytes':'#b5bd61',
'Megakaryocytes':'#e377c2',
'NK':'#8c564b'}
dict_adata = si.read_embedding()
adata_C = dict_adata['C'] # embeddings for cells
adata_G = dict_adata['G'] # embeddings for genes
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CG[adata_C.obs_names,:].obs['celltype'].copy()
adata_C
[28]:
AnnData object with n_obs × n_vars = 2700 × 50
obs: 'celltype'
[29]:
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
si.pl.umap(adata_C,color=['celltype'],
dict_palette={'celltype': palette_celltype},
fig_size=(6,4),
drawing_order='random')
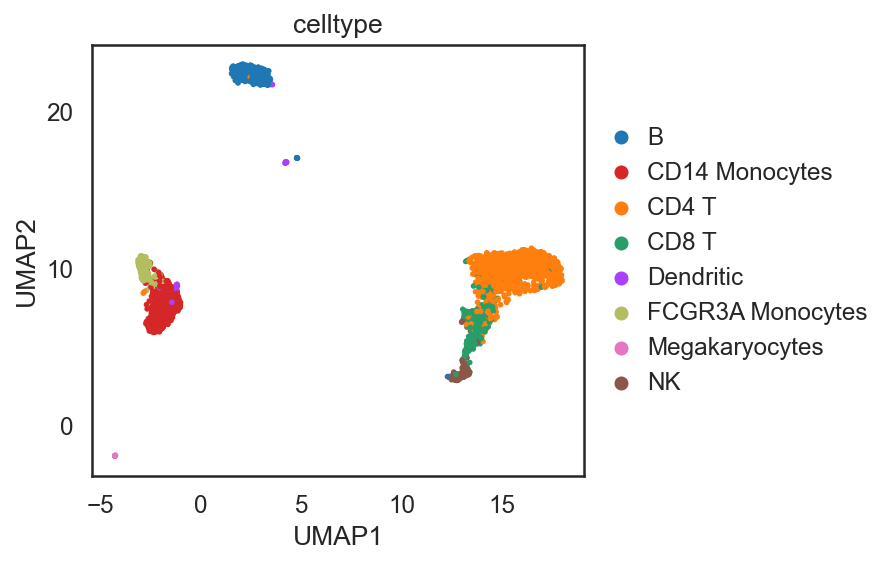
[ ]:
Using edge weights (raw gene expression) - only variable genes
[30]:
si.tl.gen_graph(list_CG=[adata_CG],
copy=False,
use_highly_variable=True,
layer=None,
add_edge_weights=True,
dirname='graph3')
relation0: source: C, destination: G
#edges: 509430
Total number of edges: 509430
Writing graph file "pbg_graph.txt" to "result_simba_edge_weights/pbg/graph3" ...
Finished.
[31]:
si.settings.pbg_params
[31]:
{'entity_path': 'result_simba_edge_weights/pbg/graph3/input/entity',
'edge_paths': ['result_simba_edge_weights/pbg/graph3/input/edge'],
'checkpoint_path': 'result_simba_edge_weights/pbg/graph2/model',
'entities': {'C': {'num_partitions': 1}, 'G': {'num_partitions': 1}},
'relations': [{'name': 'r0',
'lhs': 'C',
'rhs': 'G',
'operator': 'none',
'weight': 1.0}],
'dynamic_relations': False,
'dimension': 50,
'global_emb': False,
'comparator': 'dot',
'num_epochs': 10,
'workers': 4,
'num_batch_negs': 50,
'num_uniform_negs': 50,
'loss_fn': 'softmax',
'lr': 0.1,
'early_stopping': False,
'regularization_coef': 0.0,
'wd': 0.015521,
'wd_interval': 50,
'eval_fraction': 0.05,
'eval_num_batch_negs': 50,
'eval_num_uniform_negs': 50,
'checkpoint_preservation_interval': None}
[32]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
dict_config['wd_interval'] = 10 # we usually set `wd_interval` to 10 for scRNA-seq datasets for a slower but finer training
dict_config['workers'] = 4 #The number of CPUs.
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model', use_edge_weights=True)
Auto-estimated weight decay is 0.069558
`.settings.pbg_params['wd']` has been updated to 0.069558
Converting input data ...
Edge weights are being used ...
[2022-10-10 13:50:24.363063] Using the 1 relation types given in the config
[2022-10-10 13:50:24.363674] Searching for the entities in the edge files...
[2022-10-10 13:50:25.083892] Entity type C:
[2022-10-10 13:50:25.084614] - Found 2700 entities
[2022-10-10 13:50:25.085133] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:50:25.086340] - Left with 2700 entities
[2022-10-10 13:50:25.087150] - Shuffling them...
[2022-10-10 13:50:25.089147] Entity type G:
[2022-10-10 13:50:25.089771] - Found 2000 entities
[2022-10-10 13:50:25.090276] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:50:25.091244] - Left with 2000 entities
[2022-10-10 13:50:25.092002] - Shuffling them...
[2022-10-10 13:50:25.093553] Preparing counts and dictionaries for entities and relation types:
[2022-10-10 13:50:25.094980] - Writing count of entity type C and partition 0
[2022-10-10 13:50:25.097761] - Writing count of entity type G and partition 0
[2022-10-10 13:50:25.100081] Preparing edge path result_simba_edge_weights/pbg/graph3/input/edge, out of the edges found in result_simba_edge_weights/pbg/graph3/pbg_graph.txt
using fast version
[2022-10-10 13:50:25.100748] Taking the fast train!
[2022-10-10 13:50:25.579906] - Processed 100000 edges so far...
[2022-10-10 13:50:26.069180] - Processed 200000 edges so far...
[2022-10-10 13:50:26.567087] - Processed 300000 edges so far...
[2022-10-10 13:50:27.100423] - Processed 400000 edges so far...
[2022-10-10 13:50:27.582349] - Processed 500000 edges so far...
[2022-10-10 13:50:28.612344] - Processed 509430 edges in total
Starting training ...
Finished
[33]:
si.pl.pbg_metrics(fig_ncol=1)
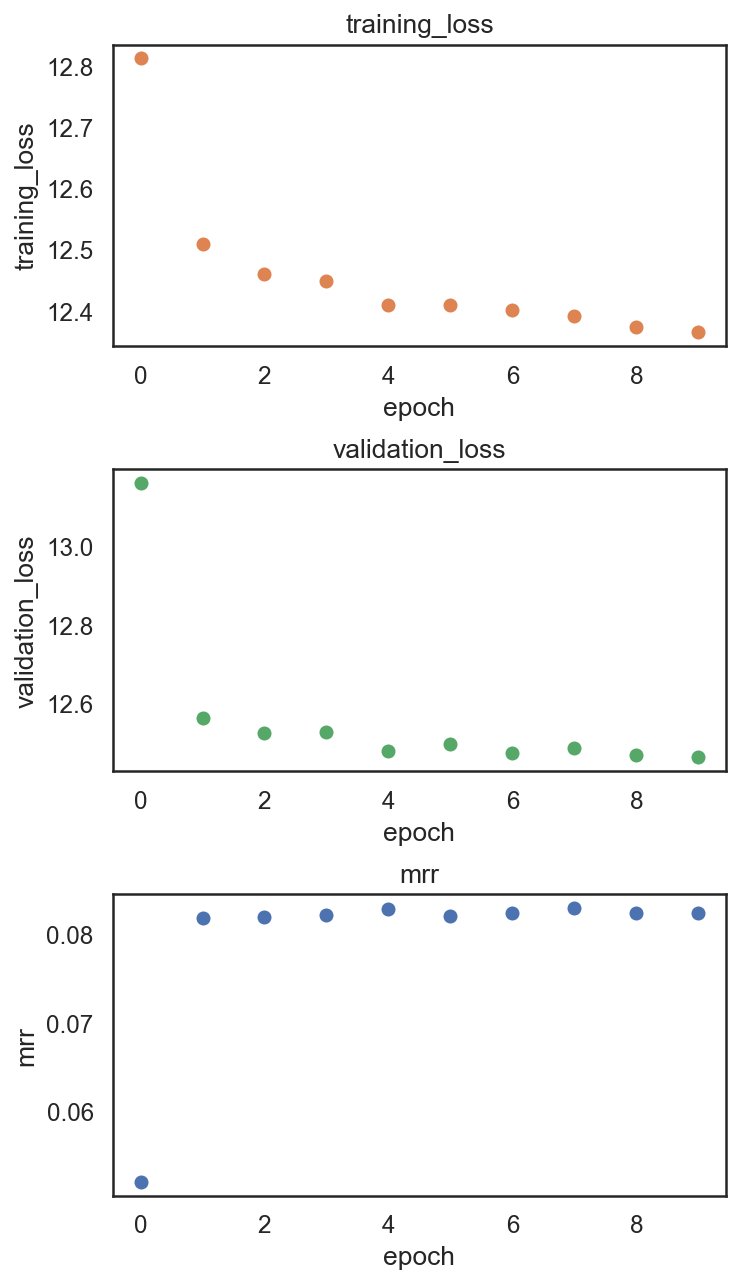
[34]:
palette_celltype={'B':'#1f77b4',
'CD4 T':'#ff7f0e',
'CD8 T':'#279e68',
'Dendritic':"#aa40fc",
'CD14 Monocytes':'#d62728',
'FCGR3A Monocytes':'#b5bd61',
'Megakaryocytes':'#e377c2',
'NK':'#8c564b'}
dict_adata = si.read_embedding()
adata_C = dict_adata['C'] # embeddings for cells
adata_G = dict_adata['G'] # embeddings for genes
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CG[adata_C.obs_names,:].obs['celltype'].copy()
adata_C
[34]:
AnnData object with n_obs × n_vars = 2700 × 50
obs: 'celltype'
[35]:
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
si.pl.umap(adata_C,color=['celltype'],
dict_palette={'celltype': palette_celltype},
fig_size=(6,4),
drawing_order='random')
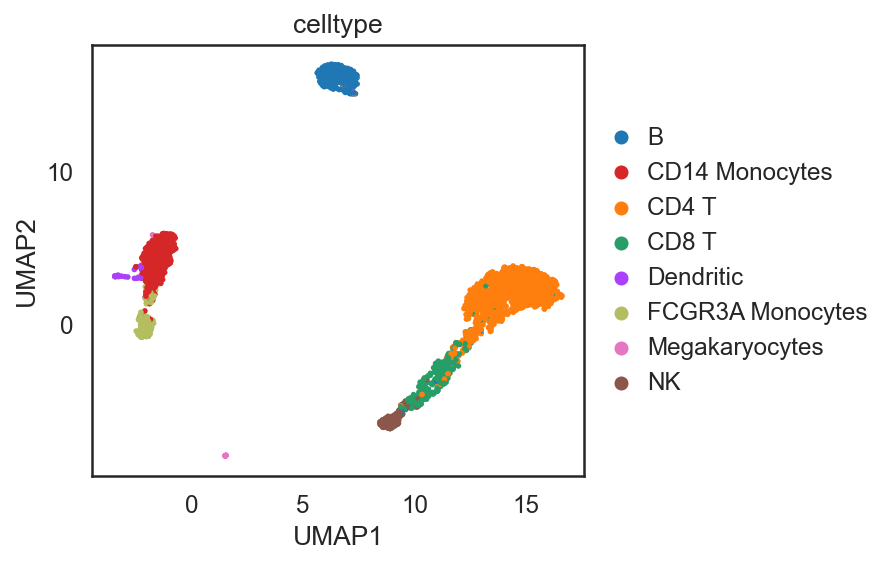
[ ]:
[ ]:
Using edge weights (discretized gene expression) - all genes
[36]:
si.tl.gen_graph(list_CG=[adata_CG],
copy=False,
use_highly_variable=False,
layer='simba',
add_edge_weights=True,
dirname='graph4')
relation0: source: C, destination: G
#edges: 2282976
Total number of edges: 2282976
Writing graph file "pbg_graph.txt" to "result_simba_edge_weights/pbg/graph4" ...
Finished.
[37]:
si.settings.pbg_params
[37]:
{'entity_path': 'result_simba_edge_weights/pbg/graph4/input/entity',
'edge_paths': ['result_simba_edge_weights/pbg/graph4/input/edge'],
'checkpoint_path': 'result_simba_edge_weights/pbg/graph3/model',
'entities': {'C': {'num_partitions': 1}, 'G': {'num_partitions': 1}},
'relations': [{'name': 'r0',
'lhs': 'C',
'rhs': 'G',
'operator': 'none',
'weight': 1.0}],
'dynamic_relations': False,
'dimension': 50,
'global_emb': False,
'comparator': 'dot',
'num_epochs': 10,
'workers': 4,
'num_batch_negs': 50,
'num_uniform_negs': 50,
'loss_fn': 'softmax',
'lr': 0.1,
'early_stopping': False,
'regularization_coef': 0.0,
'wd': 0.069558,
'wd_interval': 50,
'eval_fraction': 0.05,
'eval_num_batch_negs': 50,
'eval_num_uniform_negs': 50,
'checkpoint_preservation_interval': None}
[38]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
dict_config['wd_interval'] = 10 # we usually set `wd_interval` to 10 for scRNA-seq datasets for a slower but finer training
dict_config['workers'] = 4 #The number of CPUs.
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model', use_edge_weights=True)
Auto-estimated weight decay is 0.015521
`.settings.pbg_params['wd']` has been updated to 0.015521
Converting input data ...
Edge weights are being used ...
[2022-10-10 13:53:33.429708] Using the 1 relation types given in the config
[2022-10-10 13:53:33.430294] Searching for the entities in the edge files...
[2022-10-10 13:53:36.640605] Entity type C:
[2022-10-10 13:53:36.641172] - Found 2700 entities
[2022-10-10 13:53:36.641698] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:53:36.642450] - Left with 2700 entities
[2022-10-10 13:53:36.643278] - Shuffling them...
[2022-10-10 13:53:36.645758] Entity type G:
[2022-10-10 13:53:36.646605] - Found 13714 entities
[2022-10-10 13:53:36.647252] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:53:36.651858] - Left with 13714 entities
[2022-10-10 13:53:36.652631] - Shuffling them...
[2022-10-10 13:53:36.665425] Preparing counts and dictionaries for entities and relation types:
[2022-10-10 13:53:36.666976] - Writing count of entity type C and partition 0
[2022-10-10 13:53:36.670722] - Writing count of entity type G and partition 0
[2022-10-10 13:53:36.679475] Preparing edge path result_simba_edge_weights/pbg/graph4/input/edge, out of the edges found in result_simba_edge_weights/pbg/graph4/pbg_graph.txt
using fast version
[2022-10-10 13:53:36.680357] Taking the fast train!
[2022-10-10 13:53:37.234925] - Processed 100000 edges so far...
[2022-10-10 13:53:37.716576] - Processed 200000 edges so far...
[2022-10-10 13:53:38.195279] - Processed 300000 edges so far...
[2022-10-10 13:53:38.651373] - Processed 400000 edges so far...
[2022-10-10 13:53:39.128663] - Processed 500000 edges so far...
[2022-10-10 13:53:39.627089] - Processed 600000 edges so far...
[2022-10-10 13:53:40.143111] - Processed 700000 edges so far...
[2022-10-10 13:53:40.673668] - Processed 800000 edges so far...
[2022-10-10 13:53:41.186034] - Processed 900000 edges so far...
[2022-10-10 13:53:41.690377] - Processed 1000000 edges so far...
[2022-10-10 13:53:42.202142] - Processed 1100000 edges so far...
[2022-10-10 13:53:42.688485] - Processed 1200000 edges so far...
[2022-10-10 13:53:43.175392] - Processed 1300000 edges so far...
[2022-10-10 13:53:43.648967] - Processed 1400000 edges so far...
[2022-10-10 13:53:44.185160] - Processed 1500000 edges so far...
[2022-10-10 13:53:44.670105] - Processed 1600000 edges so far...
[2022-10-10 13:53:45.185420] - Processed 1700000 edges so far...
[2022-10-10 13:53:45.696358] - Processed 1800000 edges so far...
[2022-10-10 13:53:46.206768] - Processed 1900000 edges so far...
[2022-10-10 13:53:46.689835] - Processed 2000000 edges so far...
[2022-10-10 13:53:47.217289] - Processed 2100000 edges so far...
[2022-10-10 13:53:47.732357] - Processed 2200000 edges so far...
[2022-10-10 13:53:51.257237] - Processed 2282976 edges in total
Starting training ...
Finished
[39]:
si.pl.pbg_metrics(fig_ncol=1)
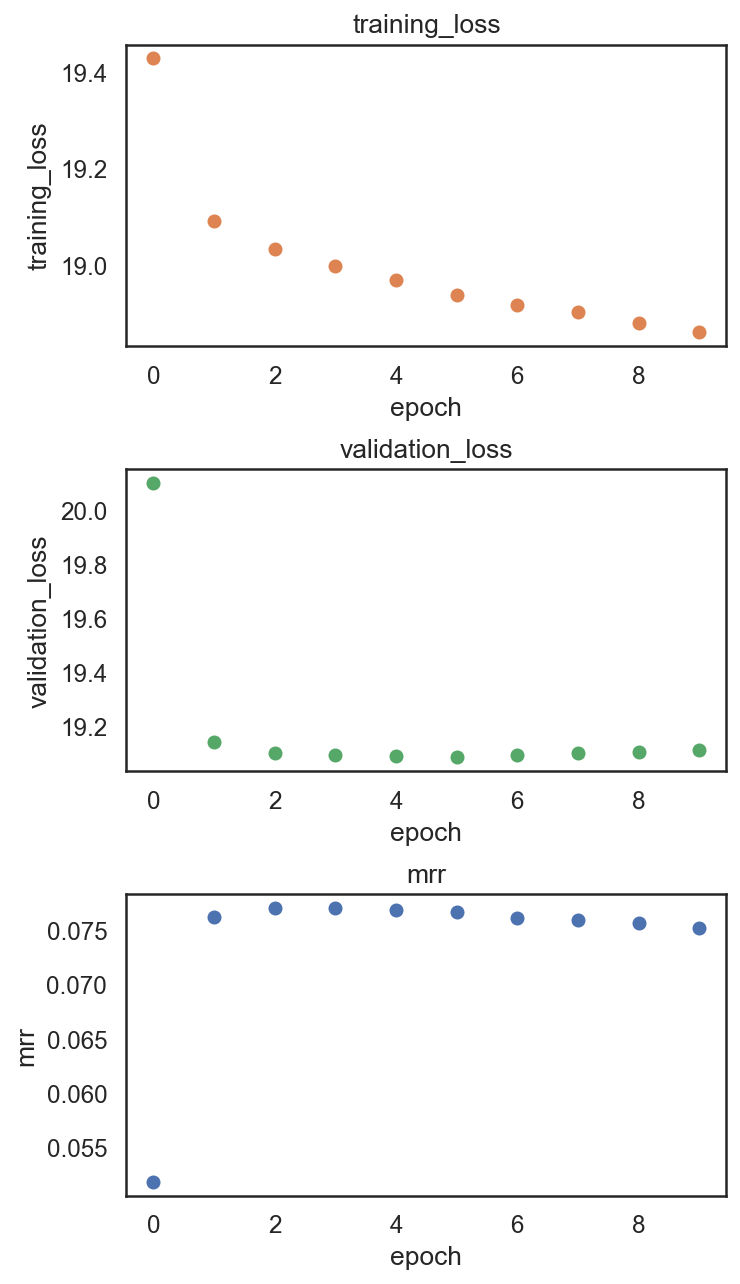
[40]:
palette_celltype={'B':'#1f77b4',
'CD4 T':'#ff7f0e',
'CD8 T':'#279e68',
'Dendritic':"#aa40fc",
'CD14 Monocytes':'#d62728',
'FCGR3A Monocytes':'#b5bd61',
'Megakaryocytes':'#e377c2',
'NK':'#8c564b'}
dict_adata = si.read_embedding()
adata_C = dict_adata['C'] # embeddings for cells
adata_G = dict_adata['G'] # embeddings for genes
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CG[adata_C.obs_names,:].obs['celltype'].copy()
adata_C
[40]:
AnnData object with n_obs × n_vars = 2700 × 50
obs: 'celltype'
[41]:
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
si.pl.umap(adata_C,color=['celltype'],
dict_palette={'celltype': palette_celltype},
fig_size=(6,4),
drawing_order='random')
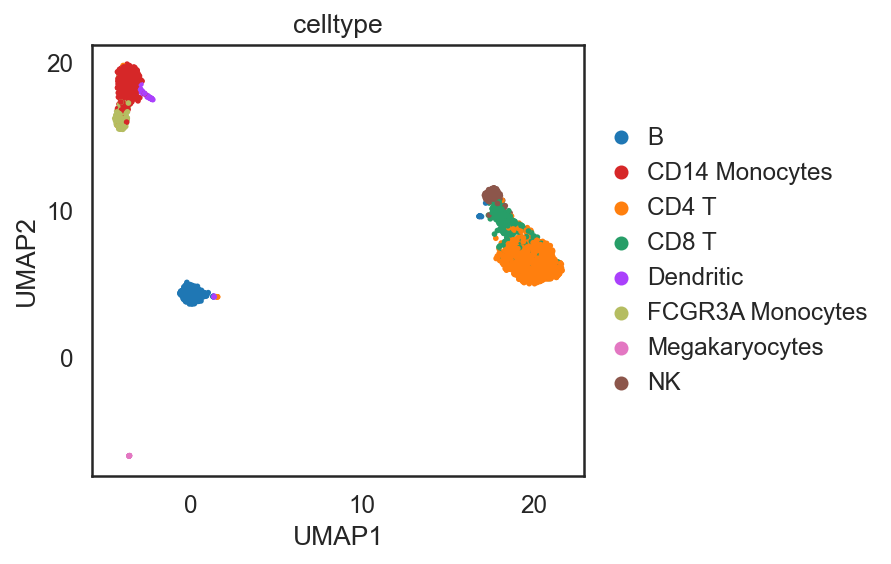
[ ]:
Using edge weights (discretized gene expression) - only variable genes
[43]:
si.tl.gen_graph(list_CG=[adata_CG],
copy=False,
use_highly_variable=True,
layer='simba',
add_edge_weights=True,
dirname='graph5')
relation0: source: C, destination: G
#edges: 509430
Total number of edges: 509430
Writing graph file "pbg_graph.txt" to "result_simba_edge_weights/pbg/graph5" ...
Finished.
[44]:
si.settings.pbg_params
[44]:
{'entity_path': 'result_simba_edge_weights/pbg/graph5/input/entity',
'edge_paths': ['result_simba_edge_weights/pbg/graph5/input/edge'],
'checkpoint_path': 'result_simba_edge_weights/pbg/graph4/model',
'entities': {'C': {'num_partitions': 1}, 'G': {'num_partitions': 1}},
'relations': [{'name': 'r0',
'lhs': 'C',
'rhs': 'G',
'operator': 'none',
'weight': 1.0}],
'dynamic_relations': False,
'dimension': 50,
'global_emb': False,
'comparator': 'dot',
'num_epochs': 10,
'workers': 4,
'num_batch_negs': 50,
'num_uniform_negs': 50,
'loss_fn': 'softmax',
'lr': 0.1,
'early_stopping': False,
'regularization_coef': 0.0,
'wd': 0.015521,
'wd_interval': 50,
'eval_fraction': 0.05,
'eval_num_batch_negs': 50,
'eval_num_uniform_negs': 50,
'checkpoint_preservation_interval': None}
[45]:
# modify parameters
dict_config = si.settings.pbg_params.copy()
dict_config['wd_interval'] = 10 # we usually set `wd_interval` to 10 for scRNA-seq datasets for a slower but finer training
dict_config['workers'] = 4 #The number of CPUs.
## start training
si.tl.pbg_train(pbg_params = dict_config, auto_wd=True, save_wd=True, output='model', use_edge_weights=True)
Auto-estimated weight decay is 0.069558
`.settings.pbg_params['wd']` has been updated to 0.069558
Converting input data ...
Edge weights are being used ...
[2022-10-10 13:57:37.849447] Using the 1 relation types given in the config
[2022-10-10 13:57:37.849988] Searching for the entities in the edge files...
[2022-10-10 13:57:38.531957] Entity type C:
[2022-10-10 13:57:38.532530] - Found 2700 entities
[2022-10-10 13:57:38.532959] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:57:38.533803] - Left with 2700 entities
[2022-10-10 13:57:38.534463] - Shuffling them...
[2022-10-10 13:57:38.536572] Entity type G:
[2022-10-10 13:57:38.537305] - Found 2000 entities
[2022-10-10 13:57:38.537740] - Removing the ones with fewer than 1 occurrences...
[2022-10-10 13:57:38.538544] - Left with 2000 entities
[2022-10-10 13:57:38.539200] - Shuffling them...
[2022-10-10 13:57:38.540654] Preparing counts and dictionaries for entities and relation types:
[2022-10-10 13:57:38.542192] - Writing count of entity type C and partition 0
[2022-10-10 13:57:38.545274] - Writing count of entity type G and partition 0
[2022-10-10 13:57:38.547926] Preparing edge path result_simba_edge_weights/pbg/graph5/input/edge, out of the edges found in result_simba_edge_weights/pbg/graph5/pbg_graph.txt
using fast version
[2022-10-10 13:57:38.548590] Taking the fast train!
[2022-10-10 13:57:39.004769] - Processed 100000 edges so far...
[2022-10-10 13:57:39.472928] - Processed 200000 edges so far...
[2022-10-10 13:57:39.980631] - Processed 300000 edges so far...
[2022-10-10 13:57:40.442172] - Processed 400000 edges so far...
[2022-10-10 13:57:40.914835] - Processed 500000 edges so far...
[2022-10-10 13:57:41.500177] - Processed 509430 edges in total
Starting training ...
Finished
[46]:
si.pl.pbg_metrics(fig_ncol=1)
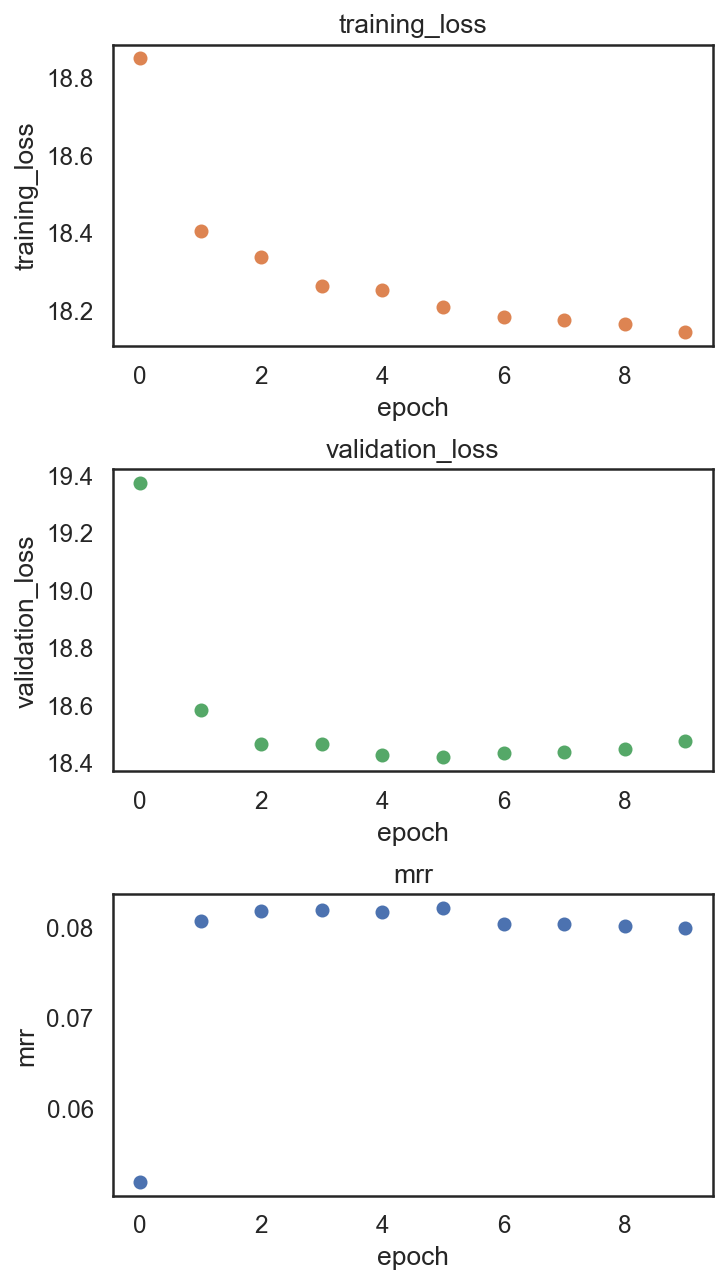
[47]:
palette_celltype={'B':'#1f77b4',
'CD4 T':'#ff7f0e',
'CD8 T':'#279e68',
'Dendritic':"#aa40fc",
'CD14 Monocytes':'#d62728',
'FCGR3A Monocytes':'#b5bd61',
'Megakaryocytes':'#e377c2',
'NK':'#8c564b'}
dict_adata = si.read_embedding()
adata_C = dict_adata['C'] # embeddings for cells
adata_G = dict_adata['G'] # embeddings for genes
## Add annotation of celltypes (optional)
adata_C.obs['celltype'] = adata_CG[adata_C.obs_names,:].obs['celltype'].copy()
adata_C
[47]:
AnnData object with n_obs × n_vars = 2700 × 50
obs: 'celltype'
[48]:
si.tl.umap(adata_C,n_neighbors=15,n_components=2)
si.pl.umap(adata_C,color=['celltype'],
dict_palette={'celltype': palette_celltype},
fig_size=(6,4),
drawing_order='random')